lincRNAs act in the circuitry controlling pluripotency and differentiation - PubMed (original) (raw)
. 2011 Aug 28;477(7364):295-300.
doi: 10.1038/nature10398.
Julie Donaghey, Bryce W Carey, Manuel Garber, Jennifer K Grenier, Glen Munson, Geneva Young, Anne Bergstrom Lucas, Robert Ach, Laurakay Bruhn, Xiaoping Yang, Ido Amit, Alexander Meissner, Aviv Regev, John L Rinn, David E Root, Eric S Lander
Affiliations
- PMID: 21874018
- PMCID: PMC3175327
- DOI: 10.1038/nature10398
lincRNAs act in the circuitry controlling pluripotency and differentiation
Mitchell Guttman et al. Nature. 2011.
Abstract
Although thousands of large intergenic non-coding RNAs (lincRNAs) have been identified in mammals, few have been functionally characterized, leading to debate about their biological role. To address this, we performed loss-of-function studies on most lincRNAs expressed in mouse embryonic stem (ES) cells and characterized the effects on gene expression. Here we show that knockdown of lincRNAs has major consequences on gene expression patterns, comparable to knockdown of well-known ES cell regulators. Notably, lincRNAs primarily affect gene expression in trans. Knockdown of dozens of lincRNAs causes either exit from the pluripotent state or upregulation of lineage commitment programs. We integrate lincRNAs into the molecular circuitry of ES cells and show that lincRNA genes are regulated by key transcription factors and that lincRNA transcripts bind to multiple chromatin regulatory proteins to affect shared gene expression programs. Together, the results demonstrate that lincRNAs have key roles in the circuitry controlling ES cell state.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. Functional effects of lincRNAs
(a) A schematic of lincRNA perturbation experiments. ESCs are infected with shRNAs, knockdown level is computed, the best hairpin is selected and profiled on expression arrays, and differential gene expression is computed relative to negative control hairpins. (b) Example of a lincRNA knockdown. Top: Genomic locus containing the lincRNA. Bottom: Heatmap of the 95 genes affected by knockdown of the lincRNA, expression for control hairpins (red line) and expression for lincRNA hairpins (blue line) are shown. (c) Distribution of number of affected genes upon knockdown of 147 lincRNAs (blue) and 40 well-known ESC regulatory proteins (red). Points corresponding to five specific ESC regulatory proteins are marked.
Figure 2. lincRNAs are critical for the maintenance of pluripotency
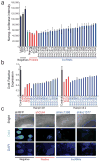
(a) Activity from a Nanog promoter driving luciferase, following treatment with control hairpins (black) or hairpins targeting luciferase (green), selected protein-coding regulators (red), and lincRNAs (blue). (b) Relative mRNA expression levels of Oct4 following knockdown of selected protein-coding (red) and lincRNA (blue) genes affecting Nanog–luciferase levels. The best hairpin (black line) and second best hairpin (grey line) are shown. All knockdowns are significant with a p-value<0.01. Error bars represent standard error (n=4). (c) Morphology of ESCs and immunofluorescence staining of Oct4 for a negative control hairpin (black line), and hairpins targeting Oct4 (red line), and two lincRNAs (blue line). The first row shows bright field images, the second row shows immunofluorescence staining of the Oct4 protein, and the third row shows DAPI staining of the nuclei.
Figure 3. lincRNAs repress specific differentiation lineages
(a) Expression changes for each lincRNA compared to gene expression of five differentiation patterns. Each box shows significant positive association (red, FDR<0.01) for Oct4 and Nanog (left) and for lincRNAs (right). (b) Expression changes upon knockdown of Oct4 and Nanog (black bars) and representative lincRNAs (grey bars) for five lineage marker genes. The expression changes (FDR<0.05) are displayed on a log scale as the _t_-statistic compared to a panel of negative control hairpins.
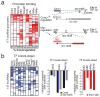
Figure 4. lincRNAs are direct regulatory targets of the ESC transcriptional circuitry
(a) A heatmap representing ChIP-Seq enrichments for 9 transcription factors (columns) at lincRNA promoters (rows). The percentage of bound lincRNAs downregulated upon knock-down of the TF are indicated in boxes (‘na’ were not measured). Right: Examples of lincRNAs from two clusters (‘core regulated’ and ‘myc regulated’) showing their genomic neighbourhood and TF binding. (b) A heatmap representing changes in lincRNA expression (rows) following knockdown of 11 TFs (columns). Middle: Effect of knockdown of Sox2, Oct4 and Nanog on expression levels of linc1405 (gray) and Oct4 (black). Right: Effect of knockdown of Klf2, Klf4, nMyc, and Esrrb on expression levels of linc1428.
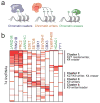
Figure 5. lincRNAs physically interact with chromatin regulatory proteins
(a) A schematic of the classes of chromatin regulators profiled: ‘readers’ (blue), ‘writers’ (orange), and ‘erasers’ (green). (b) A heatmap showing the enrichment of 74 lincRNAs (rows) for one of 12 chromatin regulatory complexes (columns). The names are color-coded by chromatin-regulatory mechanism. Major clusters are indicated by vertical lines with a description of the chromatin components.
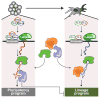
Figure 6. A model for lincRNA integration into the molecular circuitry of the cell
ESC-specific transcription factors (such as Oct4, Sox2, and Nanog) bind to the promoter of a lincRNA gene and drive its transcription. The lincRNA binds to ubiquitous regulatory proteins, giving rise to cell-type specific RNA–protein complexes. Through different combinations of protein interactions, the lincRNA–protein complex can give rise to unique transcriptional programs. Right: A similar process may also work in other cell types with specific transcription factors regulating lincRNAs, creating cell-type–specific RNA–protein complexes and regulating cell-type–specific expression programs.
Comment in
- Noncoding RNA scaffolds in pluripotency.
Mondal T, Kanduri C. Mondal T, et al. Circ Res. 2012 Apr 27;110(9):1162-5. doi: 10.1161/RES.0b013e318257c489. Circ Res. 2012. PMID: 22539754 No abstract available.
Similar articles
- Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression.
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Khalil AM, et al. Proc Natl Acad Sci U S A. 2009 Jul 14;106(28):11667-72. doi: 10.1073/pnas.0904715106. Epub 2009 Jul 1. Proc Natl Acad Sci U S A. 2009. PMID: 19571010 Free PMC article. - Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells.
Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Loewer S, et al. Nat Genet. 2010 Dec;42(12):1113-7. doi: 10.1038/ng.710. Epub 2010 Nov 7. Nat Genet. 2010. PMID: 21057500 Free PMC article. - Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation.
Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Dinger ME, et al. Genome Res. 2008 Sep;18(9):1433-45. doi: 10.1101/gr.078378.108. Epub 2008 Jun 18. Genome Res. 2008. PMID: 18562676 Free PMC article. - Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells.
Ghosal S, Das S, Chakrabarti J. Ghosal S, et al. Stem Cells Dev. 2013 Aug 15;22(16):2240-53. doi: 10.1089/scd.2013.0014. Epub 2013 May 14. Stem Cells Dev. 2013. PMID: 23528033 Free PMC article. Review. - Chromatin modifiers and remodellers: regulators of cellular differentiation.
Chen T, Dent SY. Chen T, et al. Nat Rev Genet. 2014 Feb;15(2):93-106. doi: 10.1038/nrg3607. Epub 2013 Dec 24. Nat Rev Genet. 2014. PMID: 24366184 Free PMC article. Review.
Cited by
- Advances and challenges in the use of liquid biopsy in gynaecological oncology.
Zhang Y, Tian L. Zhang Y, et al. Heliyon. 2024 Oct 15;10(20):e39148. doi: 10.1016/j.heliyon.2024.e39148. eCollection 2024 Oct 30. Heliyon. 2024. PMID: 39492906 Free PMC article. Review. - Expression of HOTAIR and PTGS2 as potential biomarkers in chronic myeloid leukemia patients in Brazil.
Kubaski Benevides AP, Marin AM, Wosniaki DK, Oliveira RN, Koerich GM, Kusma BN, Munhoz EC, Zanette DL, Aoki MN. Kubaski Benevides AP, et al. Front Oncol. 2024 Oct 10;14:1443346. doi: 10.3389/fonc.2024.1443346. eCollection 2024. Front Oncol. 2024. PMID: 39450252 Free PMC article. - Identification and functional characterization of lncRNAs involved in human monocyte-to-macrophage differentiation.
Montano C, Covarrubias S, Malekos E, Katzman S, Carpenter S. Montano C, et al. RNA Biol. 2024 Jan;21(1):39-51. doi: 10.1080/15476286.2024.2417155. Epub 2024 Oct 21. RNA Biol. 2024. PMID: 39429195 Free PMC article. - Navigating the microalgal maze: a comprehensive review of recent advances and future perspectives in biological networks.
Panahi B, Khalilpour Shadbad R. Panahi B, et al. Planta. 2024 Oct 5;260(5):114. doi: 10.1007/s00425-024-04543-7. Planta. 2024. PMID: 39367989 Review. - Comprehensive investigation of long non-coding RNA HOTAIR polymorphisms and cancer risk: a current meta-analysis encompassing 96,458 participants.
Krishna BM, Garg P, Ramisetty S, Subbalakshmi AR, Kulkarni P, Salgia R, Singhal SS. Krishna BM, et al. Sci Rep. 2024 Sep 30;14(1):22670. doi: 10.1038/s41598-024-72586-7. Sci Rep. 2024. PMID: 39349529 Free PMC article.
References
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases