Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743 - PubMed (original) (raw)
. 2011 Nov 18;6(11):1244-56.
doi: 10.1021/cb200244t. Epub 2011 Sep 20.
Benjamin Janto, Josh Earl, Azad Ahmed, Fen Z Hu, Luisa Hiller, Meg Dahlgren, Rachael Kreft, Fengan Yu, Jeremy J Wolff, Hye Kyong Kweon, Michael A Christiansen, Kristina Håkansson, Robert M Williams, Garth D Ehrlich, David H Sherman
Affiliations
- PMID: 21875091
- PMCID: PMC3220770
- DOI: 10.1021/cb200244t
Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743
Christopher M Rath et al. ACS Chem Biol. 2011.
Abstract
In many macroorganisms, the ultimate source of potent biologically active natural products has remained elusive due to an inability to identify and culture the producing symbiotic microorganisms. As a model system for developing a meta-omic approach to identify and characterize natural product pathways from invertebrate-derived microbial consortia, we chose to investigate the ET-743 (Yondelis) biosynthetic pathway. This molecule is an approved anticancer agent obtained in low abundance (10(-4)-10(-5) % w/w) from the tunicate Ecteinascidia turbinata and is generated in suitable quantities for clinical use by a lengthy semisynthetic process. On the basis of structural similarities to three bacterial secondary metabolites, we hypothesized that ET-743 is the product of a marine bacterial symbiont. Using metagenomic sequencing of total DNA from the tunicate/microbial consortium, we targeted and assembled a 35 kb contig containing 25 genes that comprise the core of the NRPS biosynthetic pathway for this valuable anticancer agent. Rigorous sequence analysis based on codon usage of two large unlinked contigs suggests that Candidatus Endoecteinascidia frumentensis produces the ET-743 metabolite. Subsequent metaproteomic analysis confirmed expression of three key biosynthetic proteins. Moreover, the predicted activity of an enzyme for assembly of the tetrahydroisoquinoline core of ET-743 was verified in vitro. This work provides a foundation for direct production of the drug and new analogues through metabolic engineering. We expect that the interdisciplinary approach described is applicable to diverse host-symbiont systems that generate valuable natural products for drug discovery and development.
Figures
Figure 1
Tetrahydroisoquinoline natural products, biosynthetic pathways, and a novel workflow. a) ET-743 (1) and: saframycin A (2), saframycin Mx1 (3), and safracin (4). b) ET-743 core modular NRPS proteins (EtuA1-3) and previously characterized Sfm, Saf, and Sac NRPS biosynthetic systems. NRPS domains are: AL-acyl ligase, T-thiolation, C-condensation, A-adenylation, RE-reductive. c) The experimental workflow: Ecteinascidia turbinata in its natural environment (Cory Walter, Mote Marine Laboratory) is collected (Erich Bartels, Mote Marine Laboratory), and subjected to meta-omic analysis. ET-743 related secondary metabolites were evaluated by Liquid chromatography FTICR mass spectrometry (LC-FTICR-MS/MS), total metagenomic and 16S DNA by 454 metagenomic sequencing with contig assembly, and total metaproteome by nano-LC-FTICR-MS/MS (nLC) and nLC-Orbitrap-MS/MS.
Figure 2
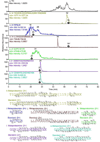
ET-743 related metabolites identified from the tunicate/microbial sample. a) LC-FTICR-MS total ion chromatogram and extracted ion chromatograms for M + H+ (b, c, f, h), and (M – H2O) + H+ (c, e, g, i) for ET-743 (1), ET-597 (19), ET-594 (21), and ET-583 (18). Y axis is in arbitrary units. All identified compounds were verified by collision induced dissociation MS/MS (Table S1).
Figure 3
Multiple sequence alignment tree. 16S rRNA gene sequences reported in previous E. turbinata analyses (26, 27) were aligned with 16S rRNA gene sequences representing the most abundant bacterial populations in our tunicate samples. A 16S rRNA gene-containing contig (00422) clusters with previously identified E. frumentensis.
Figure 4
ET-743 biosynthesis. a) Gene organization and names on the contiguous 35 kb gene cluster. Names relate to proposed function for each protein:  -NRPS with domains illustrated,
-NRPS with domains illustrated,  -DNA processing,
-DNA processing,  -fatty-acid enzymes,
-fatty-acid enzymes,  hydroxylase,
hydroxylase,  methyltransferases,
methyltransferases,  amidotransferases,
amidotransferases,  monoxygenase,
monoxygenase,  -pyruvate cassette,
-pyruvate cassette,  -regulatory enzymes,
-regulatory enzymes,  -drug transporter, **EtuU-**unknown function. b) Proposed biosynthetic pathway for ET-743. Named intermediates (characterized), enzymes (if assigned) and enzyme intermediates (thioester-bound) are shown.
-drug transporter, **EtuU-**unknown function. b) Proposed biosynthetic pathway for ET-743. Named intermediates (characterized), enzymes (if assigned) and enzyme intermediates (thioester-bound) are shown.
Figure 5
EtuA2 RE and SfmC reactions with (26). a) The proposed biochemical activity of EtuA2 RE-domain in transforming activated didepsipeptide acyl-thioester (10) to the aldehyde (11). b) The analogous reaction for SfmC is the transformation of (26) to (27) as reported by Koketsu (25). The reaction of (26) to (27) was investigated with no enzyme control (c), EtuA2 RE-domain (d), SfmC (e), and an authentic standard of (27) (f). The aldehyde-dipeptide product (27) was monitored as the Na+ adduct in positive ion mode by LC-FTICR MS with an EIC at +/−20 ppm. Inset spectra are shown over the peak elution window (g–i).
Figure 6
Synthetic peptides as authentic standards to verify metaproteomics peptide assignments. a) Total ion chromatogram for the standard peptide mixture on the LTQ-orbitrap. b–g) extracted ion chromatograms generated at +/− 0.1 m/z for each of the synthetic peptides in the mixture. Chromatograms are presented as time versus normalized intensity. Maximum intensity in each normalized total or extracted ion chromatogram is noted. *Denotes that the experimental retention time for doubly protonated tryptic LLDVGGGTAINAIALAK was obtained on a different LC system with a different gradient and column, as compared to the authentic standard. In the case of all other synthetic standard versus experimental identifications the LC system and gradient were identical, although a different column was used. ◊ denotes the elution time of the experimental MS2 spectra assigned to each of the peptides. Peptide MS2 sequence coverage for metaproteomics versus authentic standard synthetic peptides (h–m). Only b and y ion assignments are shown although other ions (e.g., a, b - H2O, b - NH3, y - H2O, and y - NH3) could also be assigned. Multiple bars indicate that a given fragment can be assigned to multiple charge states.
Similar articles
- Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743.
Schofield MM, Jain S, Porat D, Dick GJ, Sherman DH. Schofield MM, et al. Environ Microbiol. 2015 Oct;17(10):3964-75. doi: 10.1111/1462-2920.12908. Epub 2015 Jul 21. Environ Microbiol. 2015. PMID: 26013440 Free PMC article. - Development of Yondelis (trabectedin, ET-743). A semisynthetic process solves the supply problem.
Cuevas C, Francesch A. Cuevas C, et al. Nat Prod Rep. 2009 Mar;26(3):322-37. doi: 10.1039/b808331m. Epub 2009 Jan 7. Nat Prod Rep. 2009. PMID: 19240944 Review. - Biosynthesis of Tetrahydroisoquinoline Antibiotics.
Tang GL, Tang MC, Song LQ, Zhang Y. Tang GL, et al. Curr Top Med Chem. 2016;16(15):1717-26. doi: 10.2174/1568026616666151012112329. Curr Top Med Chem. 2016. PMID: 26456466 Review. - Trabectedin: Ecteinascidin 743, Ecteinascidin-743, ET 743, ET-743, NSC 684766.
[No authors listed] [No authors listed] Drugs R D. 2006;7(5):317-28. doi: 10.2165/00126839-200607050-00005. Drugs R D. 2006. PMID: 16922593 Review. - A review of trabectedin (ET-743): a unique mechanism of action.
D'Incalci M, Galmarini CM. D'Incalci M, et al. Mol Cancer Ther. 2010 Aug;9(8):2157-63. doi: 10.1158/1535-7163.MCT-10-0263. Epub 2010 Jul 20. Mol Cancer Ther. 2010. PMID: 20647340 Review.
Cited by
- Data-independent microbial metabolomics with ambient ionization mass spectrometry.
Rath CM, Yang JY, Alexandrov T, Dorrestein PC. Rath CM, et al. J Am Soc Mass Spectrom. 2013 Aug;24(8):1167-76. doi: 10.1007/s13361-013-0608-y. Epub 2013 Apr 9. J Am Soc Mass Spectrom. 2013. PMID: 23568029 Free PMC article. - Heterologous production of 4-O-demethylbarbamide, a marine cyanobacterial natural product.
Kim EJ, Lee JH, Choi H, Pereira AR, Ban YH, Yoo YJ, Kim E, Park JW, Sherman DH, Gerwick WH, Yoon YJ. Kim EJ, et al. Org Lett. 2012 Dec 7;14(23):5824-7. doi: 10.1021/ol302575h. Epub 2012 Nov 13. Org Lett. 2012. PMID: 23148802 Free PMC article. - Marine Anticancer Agents: An Overview with a Particular Focus on Their Chemical Classes.
Barreca M, Spanò V, Montalbano A, Cueto M, Díaz Marrero AR, Deniz I, Erdoğan A, Lukić Bilela L, Moulin C, Taffin-de-Givenchy E, Spriano F, Perale G, Mehiri M, Rotter A, P Thomas O, Barraja P, Gaudêncio SP, Bertoni F. Barreca M, et al. Mar Drugs. 2020 Dec 4;18(12):619. doi: 10.3390/md18120619. Mar Drugs. 2020. PMID: 33291602 Free PMC article. Review. - Genome streamlining and chemical defense in a coral reef symbiosis.
Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. Kwan JC, et al. Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):20655-60. doi: 10.1073/pnas.1213820109. Epub 2012 Nov 26. Proc Natl Acad Sci U S A. 2012. PMID: 23185008 Free PMC article. - The secret to a successful relationship: lasting chemistry between ascidians and their symbiotic bacteria.
Schmidt EW. Schmidt EW. Invertebr Biol. 2015 Mar 1;134(1):88-102. doi: 10.1111/ivb.12071. Invertebr Biol. 2015. PMID: 25937788 Free PMC article.
References
- Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. - PubMed
- Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: potent antitumor agents from the Caribbean tunicate Ecteinascidia turbinata. J. Org. Chem. 1990;55:4512–4515.
- Izbicka E, Lawrence R, Raymond E, Eckhardt G, Faircloth G, Jimeno J, Clark G, Von Hoff DD. In vitro antitumor activity of the novel marine agent, ecteinascidin-743 (ET-743, NSC-648766) against human tumors explanted from patients. Annals Oncol. 1998;9:981–987. - PubMed
- Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn KW. DNA sequence- and structure-selective alkylation of guanine N2 in the DNA minor groove by Ecteinascidin 743, a potent antitumor compound from the Caribbean tunicate Ecteinascidia turbinata. Biochem. 1996;35:13303–13309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA070375/CA/NCI NIH HHS/United States
- U01 TW007404/TW/FIC NIH HHS/United States
- R01 CA070375/CA/NCI NIH HHS/United States
- R01 CA070375-08/CA/NCI NIH HHS/United States
- C76HF00659/PHS HHS/United States
- U01 TW007404-08/TW/FIC NIH HHS/United States
- R01 DC002148/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials