Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing - PubMed (original) (raw)
Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing
Jennifer A Broderick et al. RNA. 2011 Oct.
Abstract
Small RNAs loaded into Argonaute proteins direct silencing of complementary target mRNAs. It has been proposed that multiple, imperfectly complementary small interfering RNAs or microRNAs, when bound to the 3' untranslated region of a target mRNA, function cooperatively to silence target expression. We report that, in cultured human HeLa cells and mouse embryonic fibroblasts, Argonaute1 (Ago1), Ago3, and Ago4 act cooperatively to silence both perfectly and partially complementary target RNAs bearing multiple small RNA-binding sites. Our data suggest that for Ago1, Ago3, and Ago4, multiple, adjacent small RNA-binding sites facilitate cooperative interactions that stabilize Argonaute binding. In contrast, small RNAs bound to Ago2 and pairing perfectly to an mRNA target act independently to silence expression. Noncooperative silencing by Ago2 does not require the endoribonuclease activity of the protein: A mutant Ago2 that cannot cleave its mRNA target also silences noncooperatively. We propose that Ago2 binds its targets by a mechanism fundamentally distinct from that used by the three other mammalian Argonaute proteins.
Figures
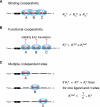
FIGURE 1.
Potential sources of cooperativity in the repression of a target mRNA by the small RNA-directed Argonaute complex, RISC. (A) Cooperative binding. RISC binding at multiple target sites increases site occupancy by mutually stabilizing subsequent binding of RISCs. (B) Cooperative function. RISC binding at multiple sites may increase the likelihood that repressive factors, such as nucleases, are recruited to the mRNA. (C) Multiple independent sites. Each RISC functions independently, so the multiple sites increase the probability of repression but do not influence each other.
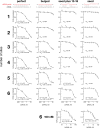
FIGURE 2.
Extent of pairing and target site number determine both efficacy and cooperativity in small RNA-directed silencing in HeLa cells. Silencing of a Renilla luciferase reporter mRNA bearing 1–6 target sites in its 3′ UTR, relative to a firefly luciferase internal control, was determined at different siRNA concentrations. Pairing between the siRNA guide (red) to the 3′ UTR sites (black) is shown at top. IC50 and Hill coefficient (nH) were calculated for each dose-response curve. Throughout this study, values are reported as mean ± standard deviation for IC50 values and nH; error bars indicate standard error for ≥12 biological replicates. The curves correspond to the concentration-dependence of silencing expected for the mean IC50 and nH values.
FIGURE 3.
siRNA validation in HeLa cells. Each siRNA was functional in silencing a reporter containing a single perfect target site. (A) Perfect siRNA. (B) Bulged siRNA. (C) siRNA with seed plus supplementary 3′ pairing (nt 13–16). (D) siRNA with only seed pairing. The curves correspond to the concentration-dependence of silencing expected for the mean IC50 and nH values (± standard deviation) calculated from three independent trials.
FIGURE 4.
Cooperative binding of RISC requires adjacent target sites in HeLa cells. Three sites spaced 19 nt apart (A) require more siRNA to achieve half-maximal silencing, compared to three adjacent sites (B), and act noncooperatively. In contrast, a perfectly matched siRNA silences a three-site reporter with sites separated by 19 nt (C) or a reporter with three adjacent sites (D) with equal efficacy and without detectable cooperativity. The three adjacent-site experiments in this figure were performed independently from those in Figure 2. A one sample, two-tailed Student's _t_-test was used to calculate the _P_-values at 95% confidence for the Hill coefficients to determine if nH was significantly different from the null hypothesis: nH = 1 (i.e., noncooperative).
FIGURE 5.
Silencing in Ago2−/− MEFs or Ago2−/− MEFs reconstituted with mouse Ago2 or catalytically inactive, mutant Ago2D669A or Ago1−/− MEFs. (A) In the absence of Ago2, silencing by a perfect site (nH = 1.6 ± 0.4; P = 0.03) is equally cooperative as a bulged site (nH = 1.8 ± 0.3; P = 0.006). (B) Mouse Ago2 expression restored noncooperative silencing by the perfect siRNA (black; nH = 1.0 ± 0.1); silencing directed by a bulged siRNA became less cooperative (red; nH = 1.5 ± 0.2; P = 0.02) than in the absence of Ago2 (red in A; nH = 1.8 ± 0.3). (C) Catalytically inactive mouse Ago2D669A likewise restored noncooperative silencing by a perfect siRNA (black; nH = 1.1 ± 0.1), but silencing by the bulged siRNA (red; nH = 1.5 ± 0.3; P = 0.04), was cooperative. (D) In the absence of Ago1, silencing by the perfect siRNA was not cooperative (black; nH = 1.1 ± 0.1), but silencing by the bulged siRNA was cooperative (red; nH = 1.7 ± 0.2; P = 0.003). A one sample, two-tailed Student's _t_-test was used to calculate the _P_-values at 95% confidence for the Hill coefficients to determine if nH was significantly different from the null hypothesis: nH = 1 (i.e., noncooperative).
FIGURE 6.
Ago1 and Ago2 protein levels in MEF cells. Ago2 was detected by Western blotting using a rabbit anti-Ago1 antibody that recognizes both mouse and human Ago1 and a rabbit anti-Ago2 antibody that recognizes both mouse and human Ago2. Ago protein levels were normalized to actin, and the level of Ago protein in wild-type MEFs was set to 1. Data are mean ± standard deviation for three trials. Inset shows representative data from a single experiment.
FIGURE 7.
In the absence of Ago2, effective silencing requires adjacent sites. (A,B) Both perfect (nH = 2.1 ± 0.3; P = 0.007) and bulged (nH = 1.5 ± 0.3; P = 0.04) adjacent sites were silenced cooperatively in the absence of Ago2. (C,D) In Ago2−/− MEFs, three target sites spaced 19 nt apart did not silence the reporter. (E,F) Expressing mouse Ago2 in the Ago2−/− MEFs allowed three distant sites to silence the reporter. (G,H) Expressing catalytically inactive, mutant Ago2D669A also allowed three distant sites to silence the reporter. (I,J) In the Ago1−/− MEFs the three distant sites silenced the reporter. A one sample, two-tailed Student's _t_-test was used to calculate the _P_-values at 95% confidence for the Hill coefficients to determine if nH was significantly different from the null hypothesis: nH = 1 (i.e., noncooperative).
Similar articles
- Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54.
Chu CY, Rana TM. Chu CY, et al. PLoS Biol. 2006 Jul;4(7):e210. doi: 10.1371/journal.pbio.0040210. PLoS Biol. 2006. PMID: 16756390 Free PMC article. - Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway.
Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Ghildiyal M, et al. RNA. 2010 Jan;16(1):43-56. doi: 10.1261/rna.1972910. Epub 2009 Nov 16. RNA. 2010. PMID: 19917635 Free PMC article. - Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins.
Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Wang B, et al. Nat Struct Mol Biol. 2009 Dec;16(12):1259-66. doi: 10.1038/nsmb.1712. Epub 2009 Nov 29. Nat Struct Mol Biol. 2009. PMID: 19946268 - From the Argonauts Mythological Sailors to the Argonautes RNA-Silencing Navigators: Their Emerging Roles in Human-Cell Pathologies.
Pantazopoulou VI, Georgiou S, Kakoulidis P, Giannakopoulou SN, Tseleni S, Stravopodis DJ, Anastasiadou E. Pantazopoulou VI, et al. Int J Mol Sci. 2020 Jun 3;21(11):4007. doi: 10.3390/ijms21114007. Int J Mol Sci. 2020. PMID: 32503341 Free PMC article. Review. - Anatomy of four human Argonaute proteins.
Nakanishi K. Nakanishi K. Nucleic Acids Res. 2022 Jul 8;50(12):6618-6638. doi: 10.1093/nar/gkac519. Nucleic Acids Res. 2022. PMID: 35736234 Free PMC article. Review.
Cited by
- Argonaute-CLIP delineates versatile, functional RNAi networks in Aedes aegypti, a major vector of human viruses.
Rozen-Gagnon K, Gu M, Luna JM, Luo JD, Yi S, Novack S, Jacobson E, Wang W, Paul MR, Scheel TKH, Carroll T, Rice CM. Rozen-Gagnon K, et al. Cell Host Microbe. 2021 May 12;29(5):834-848.e13. doi: 10.1016/j.chom.2021.03.004. Epub 2021 Mar 31. Cell Host Microbe. 2021. PMID: 33794184 Free PMC article. - Dicer partner proteins tune the length of mature miRNAs in flies and mammals.
Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Fukunaga R, et al. Cell. 2012 Oct 26;151(3):533-46. doi: 10.1016/j.cell.2012.09.027. Epub 2012 Oct 11. Cell. 2012. PMID: 23063653 Free PMC article. - Outside the limit: questioning the distance restrictions for cooperative miRNA binding sites.
Diener C, Hart M, Fecher-Trost C, Knittel J, Rheinheimer S, Meyer MR, Mayer J, Flockerzi V, Keller A, Meese E. Diener C, et al. Cell Mol Biol Lett. 2023 Jan 24;28(1):8. doi: 10.1186/s11658-023-00421-4. Cell Mol Biol Lett. 2023. PMID: 36694129 Free PMC article. - The sequence features that define efficient and specific hAGO2-dependent miRNA silencing guides.
Yan Y, Acevedo M, Mignacca L, Desjardins P, Scott N, Imane R, Quenneville J, Robitaille J, Feghaly A, Gagnon E, Ferbeyre G, Major F. Yan Y, et al. Nucleic Acids Res. 2018 Sep 19;46(16):8181-8196. doi: 10.1093/nar/gky546. Nucleic Acids Res. 2018. PMID: 30239883 Free PMC article. - MicroRNA (miRNA)-to-miRNA Regulation of Programmed Cell Death 4 (PDCD4).
Ajuyah P, Hill M, Ahadi A, Lu J, Hutvagner G, Tran N. Ajuyah P, et al. Mol Cell Biol. 2019 Aug 27;39(18):e00086-19. doi: 10.1128/MCB.00086-19. Print 2019 Sep 15. Mol Cell Biol. 2019. PMID: 31235478 Free PMC article.
References
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell 4: 625–637 - PubMed
- Ameres SL, Martinez J, Schroeder R 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 - PubMed
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM65236/GM/NIGMS NIH HHS/United States
- R01 GM065236/GM/NIGMS NIH HHS/United States
- GM62862/GM/NIGMS NIH HHS/United States
- R37 GM062862/GM/NIGMS NIH HHS/United States
- R01 NS038194/NS/NINDS NIH HHS/United States
- R01 GM062862/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials