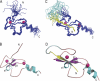Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF) - PubMed (original) (raw)
Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF)
Su Qin et al. J Biol Chem. 2011.
Abstract
MOZ (monocytic leukemic zinc-finger protein) and MORF (MOZ-related factor) are histone acetyltransferases important for HOX gene expression as well as embryo and postnatal development. They form complexes with other regulatory subunits through the scaffold proteins BRPF1/2/3 (bromodomain-PHD (plant homeodomain) finger proteins 1, 2, or 3). BRPF proteins have multiple domains, including two PHD fingers, for potential interactions with histones. Here we show that the first PHD finger of BRPF2 specifically recognizes the N-terminal tail of unmodified histone H3 (unH3) and report the solution structures of this PHD finger both free and in complex with the unH3 peptide. Structural analysis revealed that the unH3 peptide forms a third antiparallel β-strand that pairs with the PHD1 two-stranded antiparallel β-sheet. The binding specificity was determined primarily through the recognition of arginine 2 and lysine 4 of the unH3 by conserved aspartic acids of PHD1 and of threonine 6 of the unH3 by a conserved asparagine. Isothermal titration calorimetry and NMR assays showed that post-translational modifications such as H3R2me2as, H3T3ph, H3K4me, H3K4ac, and H3T6ph antagonized the interaction between histone H3 and PHD1. Furthermore, histone binding by PHD1 was important for BRPF2 to localize to the HOXA9 locus in vivo. PHD1 is highly conserved in yeast NuA3 and other histone acetyltransferase complexes, so the results reported here also shed light on the function and regulation of these complexes.
Figures
FIGURE 1.
BRPF2-PHD1 finger preferentially binds to histone H3K4me0. A, shown is a diagram of BRPF2 domain architecture. B, GST-PHD1, but not GST-PHD2 or GST alone, interacts with histones, visualized by Coomassie staining. C, ITC measurements of the binding of the BRPF2-PHD1 finger to unmodified AND mono- and trimethylated H3K4 peptides (residues 1–12) are shown. D, six superimposed 1H,15N HSQC spectra of PHD1 (0.36 m
m
) collected during titration of H3K4me0 peptide are color-coded according to the ligand. Inset, PHD1 molal ratio.
FIGURE 2.
Solution structures of BRPF2-PHD1 finger, both free and fused to unH3(1–12). Shown are the 20 lowest energy structures of the BRPF2-PHD1 finger, both free (A) and fused to unH3 (C). Ribbon representations of the lowest-energy structure of the BRPF2-PHD1 finger, both free (B) and fused to unH3 (D) are shown. Pink spheres represent Zn2+ atoms. Histone peptide and linker are shown in yellow and pale cyan, respectively.
FIGURE 3.
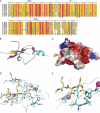
Details of the interaction of the BRPF2-PHD1 finger with the linked unH3. A, sequence alignment of PZPMs in scaffold proteins of NuA3-like histone acetyltransferase complex are shown. PHD1 zinc binding residues are indicated by blue stars or blue circles; PHD2 zinc binding residues are indicated by blue stars for blue circles; potential mononuclear Zn2+ knuckle zinc-binding residues are indicated by black boxes, and residues mutated for interaction assays in this study are indicated by red triangles. Numbering refers to the human BRPF2. The prefix Hs indicates Homo sapiens; Dr, Danio rerio; Dm, Drosophila melanogaster; Ce, C. elegans; Sc, Saccharomyces cerevisiae. The sequence alignment was performed using ClustalW, and the panel was generated by ESPript 2.2. B, shown is a ribbon representation of the complex highlighting the secondary structural elements. C, shown is electrostatic potential (isocontour value of ±76 kT/e) surface representation of the BRPF2-PHD1 bound to the H3K4me0 peptide (yellow). D, key protein-peptide polar interactions are shown in stick mode. E, H3A1- and H3T3-interacting neighborhood hydrophobic pockets are shown. D and E, the peptide and protein residues are color-coded by atom type with carbon atoms in yellow and cyan, respectively. The orientation of the peptide is the same as that in B.
FIGURE 4.
Backbone NMR relaxation data for BRPF2-PHD1, both free and bound to unH3 peptide. 15N longitudinal (T1) (A) and transversal (T2) (B) relaxation times and heteronuclear {1H}-15N NOEs (C) are represented for residues of PHD in its free form and bound to the unlabeled unH3(1–12) peptide. D, shown is a schematic representation of the BRPF2-PHD1-unH3 complex with cyan and yellow representing BRPF2-PHD1 and unH3, respectively. Residues displaying an increase of the heteronuclear NOE values upon peptide binding are represented in stick form (dirty violet).
FIGURE 5.
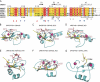
Comparison of histone recognition by the different PHD fingers. A, shown is sequence alignment of the PHD fingers of human BRPFs, AIRE, and BHC80. Secondary structure elements of free BRPF2-PHD1 and BHC80-PHD are shown. The numbering refers to the human BRPF2. The same symbols from Fig. 3_A_ are used. B–G, selected PHD-H3 complexes are shown in schematic mode, and the side chains of H3R2, H3K4, and H3T6 (H3K14 for panel G) of histone H3 and their specific interactions with the different PHD fingers are explicitly shown. For DNMT3A-ADD, only the PHD-fold portion is shown for clarity.
FIGURE 6.
Histone binding by PHD1 is important for BRPF2 to bind to the HOXA9 locus in vivo. A, 293T cells were transiently transfected with an empty vector, wild-type full-length FLAG-tagged BRPF2, or full-length BRPF2 that had either N229A/D235A or D212A/D214A point mutations. A Western blot with anti-FLAG antibodies indicated that all three constructs were expressed at high levels. Ctrl, control. B, ChIP assays of the HOXA9 gene from the transfected cells in A are shown. Ab, antibody.
Similar articles
- Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes.
Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, Champagne N, Côté J, Yang XJ. Ullah M, et al. Mol Cell Biol. 2008 Nov;28(22):6828-43. doi: 10.1128/MCB.01297-08. Epub 2008 Sep 15. Mol Cell Biol. 2008. PMID: 18794358 Free PMC article. - Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain.
Poplawski A, Hu K, Lee W, Natesan S, Peng D, Carlson S, Shi X, Balaz S, Markley JL, Glass KC. Poplawski A, et al. J Mol Biol. 2014 Apr 17;426(8):1661-76. doi: 10.1016/j.jmb.2013.12.007. Epub 2013 Dec 12. J Mol Biol. 2014. PMID: 24333487 Free PMC article. - Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin.
Ali M, Yan K, Lalonde ME, Degerny C, Rothbart SB, Strahl BD, Côté J, Yang XJ, Kutateladze TG. Ali M, et al. J Mol Biol. 2012 Dec 14;424(5):328-38. doi: 10.1016/j.jmb.2012.10.004. Epub 2012 Oct 12. J Mol Biol. 2012. PMID: 23063713 Free PMC article. - MOZ and MORF acetyltransferases: Molecular interaction, animal development and human disease.
Yang XJ. Yang XJ. Biochim Biophys Acta. 2015 Aug;1853(8):1818-26. doi: 10.1016/j.bbamcr.2015.04.014. Epub 2015 Apr 25. Biochim Biophys Acta. 2015. PMID: 25920810 Review. - Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes.
Klein BJ, Lalonde ME, Côté J, Yang XJ, Kutateladze TG. Klein BJ, et al. Epigenetics. 2014 Feb;9(2):186-93. doi: 10.4161/epi.26792. Epub 2013 Oct 29. Epigenetics. 2014. PMID: 24169304 Free PMC article. Review.
Cited by
- Structural insights into recognition of acetylated histone ligands by the BRPF1 bromodomain.
Lubula MY, Eckenroth BE, Carlson S, Poplawski A, Chruszcz M, Glass KC. Lubula MY, et al. FEBS Lett. 2014 Nov 3;588(21):3844-54. doi: 10.1016/j.febslet.2014.09.028. Epub 2014 Sep 30. FEBS Lett. 2014. PMID: 25281266 Free PMC article. - BRPF1-KAT6A/KAT6B Complex: Molecular Structure, Biological Function and Human Disease.
Zu G, Liu Y, Cao J, Zhao B, Zhang H, You L. Zu G, et al. Cancers (Basel). 2022 Aug 23;14(17):4068. doi: 10.3390/cancers14174068. Cancers (Basel). 2022. PMID: 36077605 Free PMC article. Review. - Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation.
Avvakumov N, Lalonde ME, Saksouk N, Paquet E, Glass KC, Landry AJ, Doyon Y, Cayrou C, Robitaille GA, Richard DE, Yang XJ, Kutateladze TG, Côté J. Avvakumov N, et al. Mol Cell Biol. 2012 Feb;32(3):689-703. doi: 10.1128/MCB.06455-11. Epub 2011 Dec 5. Mol Cell Biol. 2012. PMID: 22144582 Free PMC article. - Mechanistic similarities in recognition of histone tails and DNA by epigenetic readers.
Vann KR, Klein BJ, Kutateladze TG. Vann KR, et al. Curr Opin Struct Biol. 2021 Dec;71:1-6. doi: 10.1016/j.sbi.2021.04.003. Epub 2021 May 13. Curr Opin Struct Biol. 2021. PMID: 33993059 Free PMC article. Review. - 1,3-Dimethyl Benzimidazolones Are Potent, Selective Inhibitors of the BRPF1 Bromodomain.
Demont EH, Bamborough P, Chung CW, Craggs PD, Fallon D, Gordon LJ, Grandi P, Hobbs CI, Hussain J, Jones EJ, Le Gall A, Michon AM, Mitchell DJ, Prinjha RK, Roberts AD, Sheppard RJ, Watson RJ. Demont EH, et al. ACS Med Chem Lett. 2014 Sep 10;5(11):1190-5. doi: 10.1021/ml5002932. eCollection 2014 Nov 13. ACS Med Chem Lett. 2014. PMID: 25408830 Free PMC article.
References
- Lee K. K., Workman J. L. (2007) Nat. Rev. Mol. Cell Biol. 8, 284–295 - PubMed
- Yang X. J., Ullah M. (2007) Oncogene 26, 5408–5419 - PubMed
- Voss A. K., Collin C., Dixon M. P., Thomas T. (2009) Dev. Cell 17, 674–686 - PubMed
- Thomas T., Voss A. K. (2004) Front. Biosci. 9, 24–31 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases