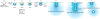Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast - PubMed (original) (raw)
Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast
Rajesh Arasada et al. Curr Biol. 2011.
Abstract
Background: Genetic analyses of budding and fission yeast identified >50 proteins that assemble at sites of clathrin-mediated endocytosis in structures called actin patches. These proteins include clathrin, clathrin-interacting proteins, actin binding proteins, and peripheral membrane proteins such as F-BAR proteins. Many questions remain regarding the interactions of these proteins, particularly the participation of F-BAR proteins in the assembly of actin filaments.
Results: Our microscopic and genetic interaction experiments on fission yeast show that F-BAR proteins Cdc15p and Bzz1p accumulate in two distinct zones on invaginating membrane tubules and interact with Myo1p and Wsp1p, nucleation-promoting factors for Arp2/3 complex. The two F-BAR proteins peak prior to movement of the actin patch and their accumulation in actin patches depends on the nucleation-promoting factors. At their peak local concentrations, we estimated the stoichiometries of the proteins in actin patches to be one Bzz1p per two Wsp1p and one Cdc15p per Myo1p. Purified Bzz1p has two SH3 domains that interact with Wsp1p and stimulate actin polymerization by Arp2/3 complex. Cells lacking either Cdc15p or Bzz1p assemble 3- to 5-fold less actin in patches (in spite of normal levels of Wsp1p, Myo1p, and Arp2/3 complex), and patches move shorter distances from the plasma membrane.
Conclusion: We propose that during clathrin-mediated endocytosis, F-BAR proteins interact with nucleation-promoting factors to stimulate Arp2/3 complex in two different zones along the invaginating tubule. We further propose that polymerization of actin filaments in these two zones contributes to membrane scission.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
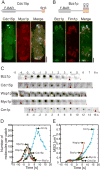
Figure 1. Quantitative analysis of actin patch dynamics
(A and B) Domain organization and localization of Cdc15p and Bzz1p in actin patches in live cells. Images are projections of 3D reconstructions. (A) Interphase cell expressing (left panel, green) mYFP-Cdc15p and (middle panel, red) mCFP-Myo1p. (Right panel) Merged image shows Cdc15p overlaps with Myo1p in many but not all actin patches because they represent different points in time. (B) Interphase cell expressing (left panel, green) Bzz1p-mEGFP and (middle panel, red) an actin binding protein fimbrin, Fim1p-mCherry. (Right panel) Merged image shows that Bzz1p overlaps with Fim1p in many but not all actin patches. Scale bar, 2 μm. (C) Montage of fluorescent micrographs (negative contrast images) of the time courses of Bzz1p-mEGFP, mEGFP-Cdc15p, mEGFP-Wsp1p, mEGFP-Myo1p and Crn1p-mEGFP accumulation and loss in actin patches. Regions of 12 ×12 pixels with an actin patch are shown at 1 s intervals for all proteins except for Crn1p-mEGFP, which is shown at 2 s intervals. Red arrows show the frame where the protein appeared and the last frame before it disappeared. Yellow arrows mark the frame where the patch started to move. Scale bar, 0.1 μm. (D) Time courses of the accumulation and loss of 5 actin patch proteins. Time zero seconds marks the initiation of patch movement. Symbols: ( , n = 20) Wsp1p; (□, n = 18) Myo1p; (▪, n = 20) Cdc15p; (
, n = 20) Wsp1p; (□, n = 18) Myo1p; (▪, n = 20) Cdc15p; ( , n=15) Bzz1p; and (
, n=15) Bzz1p; and ( , n = 30) Crn1p. (E) Time course of the mean square displacement of actin patches marked with (
, n = 30) Crn1p. (E) Time course of the mean square displacement of actin patches marked with ( , n = 20) Wsp1p, (□, n=18) Myo1p, (▪, n = 20) Cdc15p, (
, n = 20) Wsp1p, (□, n=18) Myo1p, (▪, n = 20) Cdc15p, ( , n = 15) Bzz1p and (
, n = 15) Bzz1p and ( , n = 30) Crn1p.
, n = 30) Crn1p.
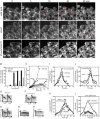
Figure 2. Effects of Bzz1p deletion or Cdc15p depletion on endocytosis and accumulation and loss of Myo1p, Wsp1p, Arp2/3 complex and actin, in actin patches
(A) Uptake of the fluorescent dye FM4-64 by endocytosis. Wild type, Δbzzl and 41xnmt1cdc15 cells were incubated on ice for 15 min to block endocytosis, exposed to 20 μM FM4-64 in YE5S at 4° C for 15 min and shifted to 25° C to restart endocytosis. The images are single optical sections through the middle of the cells at different time points. Red arrows mark fluorescent vacuolar membranes. The rows show (top) wild type cells, (middle) Δbzz1 cells and (bottom) 41xnmt1cdc15 cells depleted of Cdc15p. Scale bar 5 μm. (B) Quantitative analysis of endocytosis by scoring cells in time lapse movies that concentrated FM4-64 dye in the vacuolar membranes: (black bars) wild type cells (n= 42), (white bars) Δbzz1 cells (n= 39) and (grey bars) 41xnmt1cdc15 cells (n=24). (C - H) Movements of actin patches tagged with coronin Crn1p-mEGFP in wild type and mutant cells. (C) Mean square displacement (MSD) of the diffraction-limited spot of Crn1p-mEGFP fluorescence over time. Symbols: (o, n = 10) wild type cells; (•, n = 12) Δbzz1 cells lacking Bzz1p; (□, n = 13) 41xnmt1cdc15 cells depleted of Cdc15p; (▪, n = 12) Δwsp1 cells lacking Wsp1p; and (Δ, n = 11) Δmyo1 cells lacking Myo1p. (D - H) Kymographs of individual Crn1p-mEGFP patches from five confocal sections imaged at 1 s intervals. A 24 × 14 pixel box was sum projected into a 24 × 1-pixel vertical lane and 16 lanes (D - F) or 41 lanes (G - H) were combined horizontally to generate negative contrast kymographs. (D) Wild type cells, (E) Δbzz1 cells, (F) 41xnmt1cdc15 cells, (G) Δwsp1 cells and (H) Δmyo1 cells. Vertical red bar is 100 nm. (I - L) Time courses of the accumulation and loss of actin patch proteins at 25°C in (○) wild type cells, (•) Δbzz1 strains lacking Bzz1p and (□) 41xnmt1cdc15 cells depleted of Cdc15p. (I) mEGFP-Myo1p was expressed from the native locus and the numbers of molecules per patch were tracked over time in wild type cells (n = 21 patches), 41xnmt1cdc15 cells (n = 15) and Δ_bzz1_ cells (n = 12). (J) mEGFP-Wsp1p was expressed from the native locus and the numbers of molecules per patch were tracked over time in wild type cells (n = 12 patches), 41xnmt1cdc15 cells (n = 23) and Δbzz1 cells (n = 18). (K) The Arp2/3 complex subunit ArpC5p-mEGFP was expressed from the native locus and the numbers of molecules per patch were tracked over time in wild type cells (n = 12), 41xnmt1cdc15 cells (n = 7) and Δbzz1 cells (n = 8). (L) GFP-actin was expressed from the leu+ locus in the presence of wild type levels of native actin from a 41× nmt1 promoter in (○) wild-type cells, (•) Δ_bzz1_ cells lacking Bzz1p and (□) from a 3× nmt1 promoter in 41xnmt1cdc15 cells. The numbers of molecules per patch were tracked over time.
Figure 3. Interactions between F-BAR proteins and nucleation promoting factors in actin polymerization in actin patches
(A) Influence of Bzz1p SH3 domains on accumulation of the protein in actin patches. Time series of negative contrast fluorescence micrographs of single confocal sections at 2 s intervals: (upper row) Bzz1p-mYFP; (middle row) Bzz1pΔSH3-mYFP; and (lower row) Bzz1pΔSH3ΔSH3-mYFP cells. (B) Influence of Bzz1p SH3 domains on accumulation of GFP-actin in actin patches. GFP-actin was expressed from a 41×nmt1 promoter from the leu+ locus in (○) wild type cells, (□) bzz1_Δ_SH3 cells lacking the C-terminal SH3 domain or (•) bzz1_Δ_SH3_Δ_SH3 cells lacking both the SH3 domains. The numbers of molecules per patch were tracked over time. (C, D) Equilibrium binding of Bzz1p SH3 domains to Wsp1p-poly (p)-VCA. Conditions: soluble 1 μM Wsp1p poly (p)-VCA was incubated with a range of concentrations of glutathione beads with bound (C) GST-Bzz1pSH3SH3 (residues 521-642) or (D) GST-Bzz1pSH3 (residues 586-642) at room temperature in 25 mM Tris - HCl pH 7.4, 75 mM NaCl, 1 mM EDTA, 1 mM DTT. Beads were pelleted at 16,000 × g and the bound fraction was calculated from concentration of Wsp1p poly (p)-VCA in the supernatant quantified by SDS-PAGE, staining with Coomassie Blue and densitometry. Equilibrium dissociation constants (Kd) were determined by fitting data to binding isotherms (solid lines). (E-F) Effects of Bzz1p SH3 domains on the nucleation promoting activity of Wsp1p. Time course of the polymerization of actin measured by the fluorescence of pyrenyl-actin. Conditions: all samples contained 4 μM actin (10% pyrenyl-actin) and 50 nM S. pombe Arp2/3 complex in KMEI buffer. (E) Dependence on nucleation promoting factors: (○) no additions; (•) 500 nM Wsp1p-poly (p)-VCA; (□) 500 nM GST-Wsp1p-poly (p)-VCA; (▪) 500 nM Wsp1p-poly (p)-VCA and 1 μM GST-Bzz1pSH3. (F) Dependence of the numbers of actin filament barbed ends created by 4 μM actin (10% pyrenyl-actin), 50 nM Arp2/3 complex and 500 nM Wsp1p-poly (p)-VCA on the concentration of (□) GST-Bzz1pSH3 or (○) Bzz1pSH3. (G) Dependence of Cdc15p-mEGFP targeting to actin patches on tail domains of Myo1p. Time series of negative contrast fluorescence micrographs at 1 s intervals of single confocal planes through cells depending on Cdc15p-mEGFP and with Myo1p lacking domains: (upper panel) myo1ΔA lacking the acidic motif; (middle panel) myo1Δ3A lacking tail homology domain 3 and acidic motif; and (lower panel) myo1Δ23A lacking tail homology domains 2 and 3 and the acidic motif. Red arrowheads indicate Cdc15p-mEGFP in patches. Scale bar 1 μm.
Figure 4. Hypothesis for the contributions of F-BAR proteins Cdc15p and Bzz1p to endocytosis
Eight time points in the life of an actin patch with time zero defined as the onset of movement of patch proteins away from the plasma membrane. The plasma membrane is a black line, clathrin is grey, nucleation promoting factors Wsp1p and Myo1p are yellow, Cdc15p is green, Bzz1p is red, actin filaments are blue and numbers of molecules are in parentheses. Clathrin is recruited 2 min prior to invagination. Nucleation promoting factors are recruited beginning at -10 s and peak prior to patch movement at -2 s (Figures 1C, 1D and 1E). F-BAR proteins Cdc15p and Bzz1p begin to accumulate at -5 s and peak at the onset of patch movement (Figures 1C, 1D and 1E). Recruitment of Cdc15p requires Myo1p (Figure 3G) and both the proteins remain near the cell surface. Bzz1p binds and activates Wsp1p to stimulate the assembly of branched actin filaments by Arp2/3 complex (Figures 3C - 3F). We assume that movement of Bzz1p, Wsp1p and many other patch proteins is associated with elongation of the plasma membrane tubule. We propose that expansion of branched filaments from two distinct zones of NPFs pushes the tip of the invaginating tubule away from the cell surface (2 - 6 s) (Figure 1E) contributing to scission of the coated vesicle. Both F-BAR proteins dissociate from the invaginating tubule as the vesicle moves into the cytoplasm (Figure 1D).
Similar articles
- High-speed superresolution imaging of the proteins in fission yeast clathrin-mediated endocytic actin patches.
Arasada R, Sayyad WA, Berro J, Pollard TD. Arasada R, et al. Mol Biol Cell. 2018 Feb 1;29(3):295-303. doi: 10.1091/mbc.E17-06-0415. Epub 2017 Dec 6. Mol Biol Cell. 2018. PMID: 29212877 Free PMC article. - Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp.
Lee WL, Bezanilla M, Pollard TD. Lee WL, et al. J Cell Biol. 2000 Nov 13;151(4):789-800. doi: 10.1083/jcb.151.4.789. J Cell Biol. 2000. PMID: 11076964 Free PMC article. - Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast.
Sirotkin V, Beltzner CC, Marchand JB, Pollard TD. Sirotkin V, et al. J Cell Biol. 2005 Aug 15;170(4):637-48. doi: 10.1083/jcb.200502053. Epub 2005 Aug 8. J Cell Biol. 2005. PMID: 16087707 Free PMC article. - Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization.
Aspenström P. Aspenström P. Int Rev Cell Mol Biol. 2009;272:1-31. doi: 10.1016/S1937-6448(08)01601-8. Int Rev Cell Mol Biol. 2009. PMID: 19121815 Review. - Contractile ring formation in Xenopus egg and fission yeast.
Noguchi T, Arai R, Motegi F, Nakano K, Mabuchi I. Noguchi T, et al. Cell Struct Funct. 2001 Dec;26(6):545-54. doi: 10.1247/csf.26.545. Cell Struct Funct. 2001. PMID: 11942608 Review.
Cited by
- Use of a fluoride channel as a new selection marker for fission yeast plasmids and application to fast genome editing with CRISPR/Cas9.
Fernandez R, Berro J. Fernandez R, et al. Yeast. 2016 Oct;33(10):549-557. doi: 10.1002/yea.3178. Epub 2016 Sep 7. Yeast. 2016. PMID: 27327046 Free PMC article. - Multiple polarity kinases inhibit phase separation of F-BAR protein Cdc15 and antagonize cytokinetic ring assembly in fission yeast.
Bhattacharjee R, Hall AR, Mangione MC, Igarashi MG, Roberts-Galbraith RH, Chen JS, Vavylonis D, Gould KL. Bhattacharjee R, et al. Elife. 2023 Feb 7;12:e83062. doi: 10.7554/eLife.83062. Elife. 2023. PMID: 36749320 Free PMC article. - Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the conserved fission yeast kinase pom1.
Kettenbach AN, Deng L, Wu Y, Baldissard S, Adamo ME, Gerber SA, Moseley JB. Kettenbach AN, et al. Mol Cell Proteomics. 2015 May;14(5):1275-87. doi: 10.1074/mcp.M114.045245. Epub 2015 Feb 26. Mol Cell Proteomics. 2015. PMID: 25720772 Free PMC article. - The Tubulation Activity of a Fission Yeast F-BAR Protein Is Dispensable for Its Function in Cytokinesis.
McDonald NA, Takizawa Y, Feoktistova A, Xu P, Ohi MD, Vander Kooi CW, Gould KL. McDonald NA, et al. Cell Rep. 2016 Jan 26;14(3):534-546. doi: 10.1016/j.celrep.2015.12.062. Epub 2016 Jan 14. Cell Rep. 2016. PMID: 26776521 Free PMC article. - Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches.
Chen Q, Pollard TD. Chen Q, et al. Curr Biol. 2013 Jul 8;23(13):1154-62. doi: 10.1016/j.cub.2013.05.005. Epub 2013 May 30. Curr Biol. 2013. PMID: 23727096 Free PMC article.
References
- Kaksonen M, Toret C, Drubin D. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. - PubMed
- Conner S, Schmid S. Regulated portals of entry into the cell. Nature. 2003;422:37–44. - PubMed
- Doherty G, McMahon H. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. - PubMed
- Kaksonen M, Toret C, Drubin D. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM-026132/GM/NIGMS NIH HHS/United States
- R37 GM026132/GM/NIGMS NIH HHS/United States
- GM-026338/GM/NIGMS NIH HHS/United States
- R01 GM026338/GM/NIGMS NIH HHS/United States
- R01 GM026132/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases