Synthetic biology: integrated gene circuits - PubMed (original) (raw)
Review
Synthetic biology: integrated gene circuits
Nagarajan Nandagopal et al. Science. 2011.
Abstract
A major goal of synthetic biology is to develop a deeper understanding of biological design principles from the bottom up, by building circuits and studying their behavior in cells. Investigators initially sought to design circuits "from scratch" that functioned as independently as possible from the underlying cellular system. More recently, researchers have begun to develop a new generation of synthetic circuits that integrate more closely with endogenous cellular processes. These approaches are providing fundamental insights into the regulatory architecture, dynamics, and evolution of genetic circuits and enabling new levels of control across diverse biological systems.
Figures
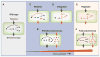
Fig. 1
A continuum of synthetic biology. Wild-type cells (A) can be subject to two basic types of synthetic manipulation. (B) Autonomous synthetic circuits, consisting of ectopic components, may be introduced into the cell. Such circuits process inputs and implement functions (orange arrows) separate from the endogenous circuitry (black). However, unknown interactions with the host cell may affect their function (purple arrows). (C) An alternative is to rewire (orange lines) the endogenous circuits themselves to have new connectivity. (D) Extending this line of synthetic manipulation, synthetic circuits could be integrated into appropriately rewired endogenous circuitry to act as sensors and to implement additional functionality. Ultimate goals of this program are to be able to design and construct (E) synthetic circuits that can functionally replace endogenous circuits or (F) fully autonomous circuits that operate independently of the cellular mileu.
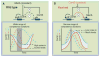
Fig. 2
Rewiring an endogenous gene circuit. (A) (Top) Part of the natural competence circuit from B. subtilis. The MecA protease adaptor (assumed to be constant) degrades ComK; ComS inhibits this degradation and thus is an indirect activator. ComK indirectly represses ComS. (Bottom) Exit from competence depends on returning to low ComS levels. Noise in ComS (white region between red and blue curves, representing the extremes of the distribution of ComS profiles in a population) is significant at such low levels. The resulting distribution in ComK curves (red dashed and blue dashed)—and thus competence durations (vertical gray bar)—is wide. (B) (Top) The rewired competence circuit: Here the activation and repression loops have been switched. Competence exit occurs when MecA levels reach a high threshold. (Bottom) The resulting distribution of competence curves is narrow because variability in MecA is relatively low at high MecA concentrations.
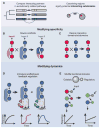
Fig. 3
Diversifying signaling pathways through rewiring. (A) Sequence correlations among proteins from a large family (left, SCA, also see text) can be used to identify interacting subdomains (right). Interaction specificity can be altered by rewiring scaffolds (B) or by shuffling specificity-determining domains or subdomains (C). The dynamic behavior of a pathway can be modified by (D) introducing new autoregulatory connections or by (E) altering regulation directly at the protein level by generating new combinations of catalytic and regulatory protein domains.
Fig. 4
An integrated synthetic circuit controls development and population dynamics. (A) In Drosophila, the synthetic Medea element (top) maternally expresses an miRNA (red) that silences a maternally expressed gene whose product is essential for embryogenesis (left column). Eggs from female flies mutant for this gene do not hatch (middle column). The Medea element also contains a rescue gene that is expressed only in the early embryo. The Medea element may also accommodate a cargo gene that is expressed in Medea progeny (right column). (B) Progeny of female _Medea_-positive flies will only survive if they receive the Medea element from either parent. (C) This super-Mendelian inheritance pattern can efficiently drive Medea into populations.
Similar articles
- Synthetic circuits, devices and modules.
Zhang H, Jiang T. Zhang H, et al. Protein Cell. 2010 Nov;1(11):974-8. doi: 10.1007/s13238-010-0133-8. Epub 2010 Dec 10. Protein Cell. 2010. PMID: 21153514 Free PMC article. Review. - Bottom-up approaches in synthetic biology and biomaterials for tissue engineering applications.
Weisenberger MS, Deans TL. Weisenberger MS, et al. J Ind Microbiol Biotechnol. 2018 Jul;45(7):599-614. doi: 10.1007/s10295-018-2027-3. Epub 2018 Mar 19. J Ind Microbiol Biotechnol. 2018. PMID: 29552703 Free PMC article. - Synthetic biology: applying biological circuits beyond novel therapies.
Dobrin A, Saxena P, Fussenegger M. Dobrin A, et al. Integr Biol (Camb). 2016 Apr 18;8(4):409-30. doi: 10.1039/c5ib00263j. Epub 2015 Dec 24. Integr Biol (Camb). 2016. PMID: 26705548 Review. - Synthetic gene networks in mammalian cells.
Weber W, Fussenegger M. Weber W, et al. Curr Opin Biotechnol. 2010 Oct;21(5):690-6. doi: 10.1016/j.copbio.2010.07.006. Epub 2010 Aug 4. Curr Opin Biotechnol. 2010. PMID: 20691580 Review. - Synthetic analog and digital circuits for cellular computation and memory.
Purcell O, Lu TK. Purcell O, et al. Curr Opin Biotechnol. 2014 Oct;29:146-55. doi: 10.1016/j.copbio.2014.04.009. Epub 2014 May 8. Curr Opin Biotechnol. 2014. PMID: 24794536 Free PMC article. Review.
Cited by
- Inducible suppression of global translation by overuse of rare codons.
Kobayashi H. Kobayashi H. Appl Environ Microbiol. 2015 Apr;81(7):2544-53. doi: 10.1128/AEM.03708-14. Epub 2015 Jan 30. Appl Environ Microbiol. 2015. PMID: 25636849 Free PMC article. - Recent advances in natural products exploitation in Streptomyces via synthetic biology.
Zhao Q, Wang L, Luo Y. Zhao Q, et al. Eng Life Sci. 2019 Mar 12;19(6):452-462. doi: 10.1002/elsc.201800137. eCollection 2019 Jun. Eng Life Sci. 2019. PMID: 32625022 Free PMC article. Review. - Synthetic approaches to study transcriptional networks and noise in mammalian systems.
Gregorio-Godoy P, Míguez DG. Gregorio-Godoy P, et al. IET Syst Biol. 2013 Feb;7(1):11-7. doi: 10.1049/iet-syb.2012.0026. IET Syst Biol. 2013. PMID: 23848051 Free PMC article. - Emerging biomedical applications of synthetic biology.
Weber W, Fussenegger M. Weber W, et al. Nat Rev Genet. 2011 Nov 29;13(1):21-35. doi: 10.1038/nrg3094. Nat Rev Genet. 2011. PMID: 22124480 Free PMC article. Review. - Evolution of an inhibitory RNA aptamer against T7 RNA polymerase.
Ohuchi S, Mori Y, Nakamura Y. Ohuchi S, et al. FEBS Open Bio. 2012 Jul 20;2:203-7. doi: 10.1016/j.fob.2012.07.004. Print 2012. FEBS Open Bio. 2012. PMID: 23650601 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- P50 GM068763/GM/NIGMS NIH HHS/United States
- 5R01GM079771/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P50GM068763/GM/NIGMS NIH HHS/United States
- R01 GM079771/GM/NIGMS NIH HHS/United States
- 5R01GM086793/GM/NIGMS NIH HHS/United States
- R01 GM086793/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources