A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles - PubMed (original) (raw)
A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles
Sebastian A Wagner et al. Mol Cell Proteomics. 2011 Oct.
Abstract
Post-translational modification of proteins by ubiquitin is a fundamentally important regulatory mechanism. However, proteome-wide analysis of endogenous ubiquitylation remains a challenging task, and almost always has relied on cells expressing affinity tagged ubiquitin. Here we combine single-step immunoenrichment of ubiquitylated peptides with peptide fractionation and high-resolution mass spectrometry to investigate endogenous ubiquitylation sites. We precisely map 11,054 endogenous putative ubiquitylation sites (diglycine-modified lysines) on 4,273 human proteins. The presented data set covers 67% of the known ubiquitylation sites and contains 10,254 novel sites on proteins with diverse cellular functions including cell signaling, receptor endocytosis, DNA replication, DNA damage repair, and cell cycle progression. Our method enables site-specific quantification of ubiquitylation in response to cellular perturbations and is applicable to any cell type or tissue. Global quantification of ubiquitylation in cells treated with the proteasome inhibitor MG-132 discovers sites that are involved in proteasomal degradation, and suggests a nonproteasomal function for almost half of all sites. Surprisingly, ubiquitylation of about 15% of sites decreased more than twofold within four hours of MG-132 treatment, showing that inhibition of proteasomal function can dramatically reduce ubiquitylation on many sites with non-proteasomal functions. Comparison of ubiquitylation sites with acetylation sites reveals an extensive overlap between the lysine residues targeted by these two modifications. However, the crosstalk between these two post-translational modifications is significantly less frequent on sites that show increased ubiquitylation upon proteasome inhibition. Taken together, we report the largest site-specific ubiquitylation dataset in human cells, and for the first time demonstrate proteome-wide, site-specific quantification of endogenous putative ubiquitylation sites.
Figures
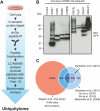
Fig. 1.
Proteome-wide mapping of endogenous ubiquitylation sites. A, Proteins from total cells lysates were digested into peptides using trypsin and ubiquitylated peptides were enriched with a monoclonal di-Gly-lysine-specific antibody. Immunoprecipitated peptides were fractionated and analyzed by mass spectrometry. Peptides with charge-state +1 and +2 were excluded from MS2 analysis to preferentially sequence ubiquitylated peptides. B, Verification of eight randomly selected proteins by Western blotting. Ubiquitylated proteins identified in our proteomic screen were affinity purified from cells expressing HA-tagged ubiquitin and ubiquitylation was confirmed by immunostaining with anti-GFP antibodies. C, Overlap of ubiquitylation sites identified in this study with sites combined from four previous MS-based ubiquitylation studies, or individually compared with the previous largest study, as indicated.
Fig. 2.
Cellular distribution and sequence properties of the ubiquitylome. A, Gene Ontology cellular compartment terms associated with ubiquitylated proteins. B, GO biological process terms associated with ubiquitylated proteins. C, Predicted protein secondary structures near ubiquitylation sites. Probabilities for different secondary structures (coil, α-helix and β-strand) of modified lysines were compared with the secondary structure probabilities of all lysines on all proteins identified in this study. D, Amino acid sequence properties of ubiquitylation sites. The heat map shows significant position-specific under- or over-representation of amino acids flanking the modification sites. E, Evolutionary conservation of ubiquitylated lysines and nonubiquitylated lysines on protein orthologs in selected eukaryotic species. Only lysine residues from ubiquitylated proteins identified in this study are compared.
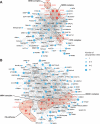
Fig. 3.
Protein interaction networks of ubiquitylated proteins. Network interaction analysis was performed using protein interaction information from the STRING database. Ubiquitylated proteins were grouped using associated GO biological process terms, and interaction subnetworks were visualized using Cytoscape. The networks show identified ubiquitylated proteins implicated in DNA replication (A) and DNA repair (B). The node size represents the number of identified ubiquitylation sites on each protein. Selected, functionally important protein core complexes are color coded in red.
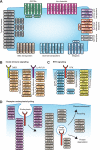
Fig. 4.
Ubiquitylation of cell surface receptors and their downstream effectors. A, The figure illustrates the functional diversity of plasma membrane expressed receptors modified by ubiquitin. B, The illustration shows ubiquitylation of key components of innate immune signaling downstream of TNF-α and IL1/toll-like receptors. C, Several RTK signaling adaptors as well as kinases involved in the mitogen activated protein -kinase and PI3-kinase signaling are ubiquitylated. D, Ubiquitylation of proteins involved in various stages of receptor trafficking.
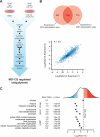
Fig. 5.
Quantification of ubiquitylation sites regulated by the proteasome inhibitor MG-132. A, Ubiquitylation sites were quantified using a SILAC method. Heavy isotope labeled MV4–11 cells were treated with MG-132 (10 μ
m
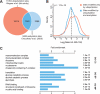
) for 4 h and light isotope labeled cells were treated with DMSO and used as control. Ubiquitylation sites were identified as described above (see Fig. 1_A_), and site-specific quantification was performed using MaxQuant. B, Experimental overlap of ubiquitylation sites identified in biological replicates. The Venn diagram shows the number of sites quantified in each experiment (upper panel). The dot plot shows the logarithmized SILAC ratios of sites quantified in both experiments. Spearman's rank correlation coefficient was calculated to determine the experimental reproducibility (lower panel). C, Treatment of cells with MG-132 results in increased, as well as decreased ubiquitylation as reflected by a broad distribution of SILAC ratios. Sites that are up- or and down-regulated are color coded in red and blue shades, respectively. The plot indicates mean ratio of ubiquitylation sites on proteins annotated with indicated PFAM, GO cellular compartment (CC) and GO biological process (BP) terms.
Fig. 6.
Site-specific crosstalk between ubiquitylation and acetylation. A, Overlap between ubiquitylation sites identified in this study and a previously published dataset of acetylation sites. B, Effect of acetylation-ubiquitylation overlap on proteasomal degradation. The SILAC ratio distribution of ubiquitylation sites after treatment with proteosome inhibitor MG-132 is compared for sites that were identified as ubiquitylated, or sites that were identified as both ubiquitylated and acetylated. C, Functional annotations of proteins modified by ubiquitylation and acetylation at the same lysine. Significantly enriched GO cellular compartment (CC), GO biological process (BP) and GO molecular function (MF) terms are shown.
Similar articles
- Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues.
Wagner SA, Beli P, Weinert BT, Schölz C, Kelstrup CD, Young C, Nielsen ML, Olsen JV, Brakebusch C, Choudhary C. Wagner SA, et al. Mol Cell Proteomics. 2012 Dec;11(12):1578-85. doi: 10.1074/mcp.M112.017905. Epub 2012 Jul 11. Mol Cell Proteomics. 2012. PMID: 22790023 Free PMC article. - Proteome-wide identification of ubiquitylation sites by conjugation of engineered lysine-less ubiquitin.
Oshikawa K, Matsumoto M, Oyamada K, Nakayama KI. Oshikawa K, et al. J Proteome Res. 2012 Feb 3;11(2):796-807. doi: 10.1021/pr200668y. Epub 2011 Nov 23. J Proteome Res. 2012. PMID: 22053931 - Ubiquitin diGLY Proteomics as an Approach to Identify and Quantify the Ubiquitin-Modified Proteome.
Fulzele A, Bennett EJ. Fulzele A, et al. Methods Mol Biol. 2018;1844:363-384. doi: 10.1007/978-1-4939-8706-1_23. Methods Mol Biol. 2018. PMID: 30242721 Free PMC article. - Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants.
Pande A, Mun BG, Khan M, Rahim W, Lee DS, Lee GM, Al Azawi TNI, Hussain A, Yun BW. Pande A, et al. Int J Mol Sci. 2022 Jan 31;23(3):1657. doi: 10.3390/ijms23031657. Int J Mol Sci. 2022. PMID: 35163578 Free PMC article. Review. - Advances in characterizing ubiquitylation sites by mass spectrometry.
Sylvestersen KB, Young C, Nielsen ML. Sylvestersen KB, et al. Curr Opin Chem Biol. 2013 Feb;17(1):49-58. doi: 10.1016/j.cbpa.2012.12.009. Epub 2013 Jan 5. Curr Opin Chem Biol. 2013. PMID: 23298953 Review.
Cited by
- Application of quantitative proteomics to the integrated analysis of the ubiquitylated and global proteomes of xenograft tumor tissues.
Thomas SN, Zhang H, Cotter RJ. Thomas SN, et al. Clin Proteomics. 2015 May 20;12(1):14. doi: 10.1186/s12014-015-9086-5. eCollection 2015. Clin Proteomics. 2015. PMID: 26019700 Free PMC article. - Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function.
Macdonald JI, Dick FA. Macdonald JI, et al. Genes Cancer. 2012 Nov;3(11-12):619-33. doi: 10.1177/1947601912473305. Genes Cancer. 2012. PMID: 23634251 Free PMC article. - Rapid identification of ubiquitination and SUMOylation target sites by microfluidic peptide array.
Zhu B, Farris TR, Milligan SL, Chen H, Zhu R, Hong A, Zhou X, Gao X, McBride JW. Zhu B, et al. Biochem Biophys Rep. 2016 Mar 1;5:430-438. doi: 10.1016/j.bbrep.2016.02.003. Biochem Biophys Rep. 2016. PMID: 27047992 Free PMC article. - The Colossus of ubiquitylation: decrypting a cellular code.
Williamson A, Werner A, Rape M. Williamson A, et al. Mol Cell. 2013 Feb 21;49(4):591-600. doi: 10.1016/j.molcel.2013.01.028. Mol Cell. 2013. PMID: 23438855 Free PMC article. - Structural insights into the activity and regulation of human Josephin-2.
Grasty KC, Weeks SD, Loll PJ. Grasty KC, et al. J Struct Biol X. 2019 Aug 21;3:100011. doi: 10.1016/j.yjsbx.2019.100011. eCollection 2019 Jul-Sep. J Struct Biol X. 2019. PMID: 32647816 Free PMC article.
References
- Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Nat. Rev. 82, 373–428 - PubMed
- Weissman A. M. (2001) Themes and variations on ubiquitylation. Mol. Cell Biol. 2, 169–178 - PubMed
- Hochstrasser M. (1995) Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Current Opinion Cell Biol. 7, 215–223 - PubMed
- Chen Z. J., Sun L. J. (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases