Genetic evidence for archaic admixture in Africa - PubMed (original) (raw)
Genetic evidence for archaic admixture in Africa
Michael F Hammer et al. Proc Natl Acad Sci U S A. 2011.
Abstract
A long-debated question concerns the fate of archaic forms of the genus Homo: did they go extinct without interbreeding with anatomically modern humans, or are their genes present in contemporary populations? This question is typically focused on the genetic contribution of archaic forms outside of Africa. Here we use DNA sequence data gathered from 61 noncoding autosomal regions in a sample of three sub-Saharan African populations (Mandenka, Biaka, and San) to test models of African archaic admixture. We use two complementary approximate-likelihood approaches and a model of human evolution that involves recent population structure, with and without gene flow from an archaic population. Extensive simulation results reject the null model of no admixture and allow us to infer that contemporary African populations contain a small proportion of genetic material (≈ 2%) that introgressed ≈ 35 kya from an archaic population that split from the ancestors of anatomically modern humans ≈ 700 kya. Three candidate regions showing deep haplotype divergence, unusual patterns of linkage disequilibrium, and small basal clade size are identified and the distributions of introgressive haplotypes surveyed in a sample of populations from across sub-Saharan Africa. One candidate locus with an unusual segment of DNA that extends for >31 kb on chromosome 4 seems to have introgressed into modern Africans from a now-extinct taxon that may have lived in central Africa. Taken together our results suggest that polymorphisms present in extant populations introgressed via relatively recent interbreeding with hominin forms that diverged from the ancestors of modern humans in the Lower-Middle Pleistocene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
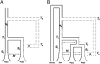
Fig. 1.
Schematic of the (A) two-population model and the (B) three-population model. Both demographic models test the fit of admixture with an archaic group (dotted lines) who split from the ancestors of modern humans at time _T_0 and a (%) of alleles introgressed into the modern gene pool at time _T_a. The dashed lines represent all possible locations where admixture could occur. Both models begin with a single population of size _N_a, followed by a population split at time _T_1, with population growth beginning at times _g_1 and _g_2, and a constant symmetric migration rate M. For B, an additional population split at time _T_2 also occurs. This model also assumes that the ancestors of the San split first from those of the Mandenka and Biaka (22).
Fig. 2.
Approximate likelihood profile based on 60 loci for time of introgression and archaic split time. A log-likelihood difference of 3.92 defines the 95% confidence region (using the χ2 approximation). Likelihood estimates at each locus have at least 10 ARGs for both ψold and ψrecent.
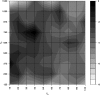
Fig. 3.
(A) Schematic of the original (filled bars) and extended sequence data (open bars) for the 4qMB179 locus. The unusual Biaka haplotype extends for ≈31.4 kb between the vertical dotted lines. (B) Recombinational landscape as inferred from HapMap Phase I data.
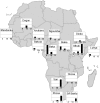
Fig. 4.
Frequency of introgressive variants within three sequenced regions in an expanded sample of ≈500 sub-Saharan Africans (
SI Materials and Methods
). The filled bar represents the frequency of a variant marking the divergent haplotype at 4qMB179 (Left), 18qMB60 (Center), and 13qMB179 (Right) in each of 14 population samples. Each horizontal line on the bar charts represents a frequency of 5%.
Similar articles
- Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies.
Hsieh P, Woerner AE, Wall JD, Lachance J, Tishkoff SA, Gutenkunst RN, Hammer MF. Hsieh P, et al. Genome Res. 2016 Mar;26(3):291-300. doi: 10.1101/gr.196634.115. Epub 2016 Feb 17. Genome Res. 2016. PMID: 26888264 Free PMC article. - Identification of African-Specific Admixture between Modern and Archaic Humans.
Wall JD, Ratan A, Stawiski E; GenomeAsia 100K Consortium. Wall JD, et al. Am J Hum Genet. 2019 Dec 5;105(6):1254-1261. doi: 10.1016/j.ajhg.2019.11.005. Am J Hum Genet. 2019. PMID: 31809748 Free PMC article. - Models of archaic admixture and recent history from two-locus statistics.
Ragsdale AP, Gravel S. Ragsdale AP, et al. PLoS Genet. 2019 Jun 10;15(6):e1008204. doi: 10.1371/journal.pgen.1008204. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31181058 Free PMC article. - Searching for archaic contribution in Africa.
Santander C, Montinaro F, Capelli C. Santander C, et al. Ann Hum Biol. 2019 Mar;46(2):129-139. doi: 10.1080/03014460.2019.1624823. Epub 2019 Jun 26. Ann Hum Biol. 2019. PMID: 31163991 Review. - Outstanding questions in the study of archaic hominin admixture.
Wolf AB, Akey JM. Wolf AB, et al. PLoS Genet. 2018 May 31;14(5):e1007349. doi: 10.1371/journal.pgen.1007349. eCollection 2018 May. PLoS Genet. 2018. PMID: 29852022 Free PMC article. Review.
Cited by
- The Nubian Complex of Dhofar, Oman: an African middle stone age industry in Southern Arabia.
Rose JI, Usik VI, Marks AE, Hilbert YH, Galletti CS, Parton A, Geiling JM, Cerný V, Morley MW, Roberts RG. Rose JI, et al. PLoS One. 2011;6(11):e28239. doi: 10.1371/journal.pone.0028239. Epub 2011 Nov 30. PLoS One. 2011. PMID: 22140561 Free PMC article. - Ancient DNA and human history.
Slatkin M, Racimo F. Slatkin M, et al. Proc Natl Acad Sci U S A. 2016 Jun 7;113(23):6380-7. doi: 10.1073/pnas.1524306113. Epub 2016 Jun 6. Proc Natl Acad Sci U S A. 2016. PMID: 27274045 Free PMC article. - Genetics and Material Culture Support Repeated Expansions into Paleolithic Eurasia from a Population Hub Out of Africa.
Vallini L, Marciani G, Aneli S, Bortolini E, Benazzi S, Pievani T, Pagani L. Vallini L, et al. Genome Biol Evol. 2022 Apr 10;14(4):evac045. doi: 10.1093/gbe/evac045. Genome Biol Evol. 2022. PMID: 35445261 Free PMC article. - Genetic variation and adaptation in Africa: implications for human evolution and disease.
Gomez F, Hirbo J, Tishkoff SA. Gomez F, et al. Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a008524. doi: 10.1101/cshperspect.a008524. Cold Spring Harb Perspect Biol. 2014. PMID: 24984772 Free PMC article. Review. - A Hominin Femur with Archaic Affinities from the Late Pleistocene of Southwest China.
Curnoe D, Ji X, Liu W, Bao Z, Taçon PS, Ren L. Curnoe D, et al. PLoS One. 2015 Dec 17;10(12):e0143332. doi: 10.1371/journal.pone.0143332. eCollection 2015. PLoS One. 2015. PMID: 26678851 Free PMC article.
References
- Coppa A, Grun R, Stringer C, Eggins S, Vargiu R. Newly recognized Pleistocene human teeth from Tabun Cave, Israel. J Hum Evol. 2005;49:301–315. - PubMed
- Grün R, et al. ESR and U-series analyses of enamel and dentine fragments of the Banyoles mandible. J Hum Evol. 2006;50:347–358. - PubMed
- Morwood MJ, et al. Archaeology and age of a new hominin from Flores in eastern Indonesia. Nature. 2004;431:1087–1091. - PubMed
- Stringer C. The origin and dispersal of Homo sapiens: Our current state of knowledge. In: Mellars P, Boyle K, Bar-Yosef O, Stringer C, editors. Rethinking the Human Revolution. Cambridge, UK: McDonald Institure for Archaeological Research; 2007.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources