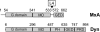Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop - PubMed (original) (raw)
Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop
Alexander von der Malsburg et al. J Biol Chem. 2011.
Abstract
The human MxA protein is an interferon-induced large GTPase with antiviral activity against a wide range of viruses, including influenza viruses. Recent structural data demonstrated that MxA oligomerizes into multimeric filamentous or ring-like structures by virtue of its stalk domain. Here, we show that negatively charged lipid membranes support MxA self-assembly. Like dynamin, MxA assembled around spherical liposomes inducing liposome tubulation. Cryo-transmission electron microscopy revealed that MxA oligomers around liposomes have a "T-bar" shape similar to dynamin. Moreover, biochemical assays indicated that the unstructured L4 loop of the MxA stalk serves as the lipid-binding moiety, and mutational analysis of L4 revealed that a stretch of four lysine residues is critical for binding. The orientation of the MxA molecule within the membrane-associated oligomer is in agreement with the proposed topology of MxA oligomers based on crystallographic data. Although oligomerization of wild-type MxA around liposomes led to the creation of helically decorated tubes similar to those formed by dynamin, this lipid interaction did not stimulate GTPase activity, in sharp contrast to the assembly-stimulated nucleotide hydrolysis observed with dynamin. Moreover, MxA readily self-assembles into rings at physiological conditions, as opposed to dynamin which self-assembles only at low salt conditions or onto lipids. Thus, the present results indicate that the oligomeric structures formed by MxA critically differ from those of dynamin.
Figures
FIGURE 1.
Domain composition of MxA and dynamin (Dyn). The schematic diagram of MxA and dynamin depicts the G domain, MD, GED, PH domain, and proline-rich domain (PRD). The numbers indicate the positions of the structural elements in the amino acid sequence. The position of the unstructured loop L4 in MxA is indicated by an arrowhead (adapted from Ref. 8).
FIGURE 2.
MxA self-assembly depends on salt and temperature. Electron microscopy images of MxA oligomers formed in vitro at different salt concentrations and temperatures are shown. MxA (250 μg/ml) was incubated in the presence of various salt concentrations in HCB for 3 h at 25 °C (a–c and e) or in HCB100 for 16 h at 4 °C (d) and then examined by negative stain TEM a–c or cryo-TEM (d–g). f and g show higher resolution cryo-TEM images of rings and filament structures. Arrowheads indicate two parallel chains of electron dense structures. Scale bars, 100 nm (a–d) and 50 nm (e–g).
FIGURE 3.
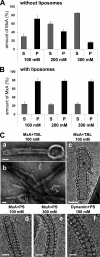
MxA binds to and tubulates liposomes by forming highly ordered arrays. A and B, sedimentation of MxA during high speed centrifugation. MxA (180 μg/ml) was diluted in HCB with the salt concentrations indicated in the absence (A) or presence (B) of TBL liposomes (1 mg/ml total brain lipids) and incubated for 20 min at 20 °C. The mixtures were separated into supernatant (S) and pellet (P) fractions by ultracentrifugation at 100,000 × g and analyzed by SDS-PAGE and Coomassie staining. The graph presents quantification of the stained protein bands of three independent experiments. Error bars, S.D. C, negative stain (a and b) and cryo-TEM images (c–e) of MxA helical arrays around liposome tubes and (f) cryo-TEM image of a dynamin-lipid tube. MxA (a–e, 250 μg/ml) or dynamin (f, 250 μg/ml) were incubated in HCB100 or HCB300 (e) for 3 h at 25 °C in the presence of liposomes generated from TBL (a–c, 500 μg/ml) or PS (d–f, 500 μg/ml). Scale bars, 100 nm (a and b) and 50 nm (c–f).
FIGURE 4.
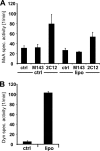
Binding of MxA to lipid bilayers does not stimulate GTPase activity. MxA (60 μg/ml) (A) or dynamin (Dyn) (160 μg/ml) (B) was diluted in HCB150 in the presence or absence (ctrl) of liposomes (1 mg/ml TBL) before starting GTP hydrolysis for 10 min at 37 °C by adding radiolabeled GTP (1 m
m
). Reaction products were analyzed by chromatography and autoradiography. MxA was preincubated for 20 min at 20 °C with MxA-specific monoclonal antibodies M134 and 2C12 (20 μg/ml). The graphs present quantification of the GTP hydrolysis of three independent experiments. Error bars, S.D.
FIGURE 5.
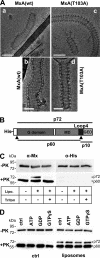
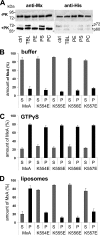
Association of wild-type and GTPase-inactive MxA with liposomes. A, cryo-TEM images of wild-type MxA (a and b) and the GTPase-inactive MxA(T103A) mutant associated with liposomes (c and d). Proteins (250 μg/ml) were incubated in HCB100 for 3 h at 25 °C in the presence of TBL liposomes (500 μg/ml). Scale bars, 100 nm (a and c) and 50 nm (b and d). B, model of the primary structure of His-tagged MxA showing the G domain, MD, the unstructured loop L4, and the GED. The two proteinase K cleavage sites (arrowheads) at amino acid positions 40 and 563 and the corresponding cleavage products p72, p60 and p10 are indicated. C, membrane binding protection of MxA from cleavage by proteinase K. MxA (180 μg/ml) was incubated in HCB150 in the absence or presence of TBL liposomes (Lipo.) (1 mg/ml) for 10 min at 20 °C. As control, the MxA-liposome mixture was incubated in buffer containing 0.5% Triton X-100 (Triton). Some samples were then treated with proteinase K (8 μg/ml) (+PK) or without (−PK) for 5 min at 20 °C. Digestion was terminated by adding SDS-sample buffer and heating for 5 min at 95 °C and was monitored by Western blot analysis using the MxA-specific antibody M143 or an antibody directed against the N-terminal His tag. D, proteinase K digestion of MxA in the presence of GTPγS. MxA (180 μg/ml) was incubated in the presence or absence (ctrl) of liposomes for 15 min at 20 °C. After the addition of the indicated nucleotides (100 μ
m
) and further incubation for 15 min at 20 °C, the samples were digested with proteinase K and analyzed by Western blotting as described for C.
FIGURE 6.
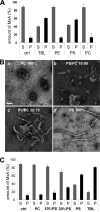
MxA preferentially binds to negatively charged phospholipids. A, MxA (180 μg/ml) was incubated in HCB300 for 20 min at 20 °C in the absence (ctrl) or presence of liposomes generated from TBL or purified phospholipids (1 mg/ml). Upon ultracentrifugation Supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE, and the intensities of the protein bands were quantified from three independent experiments. Error bars, S.D. B, tubulation of negatively charged liposomes. MxA (250 μg/ml) was incubated in HCB300 for 3 h at 25 °C in the presence of liposomes generated from PC (a), PC/PS mixtures containing 10% or 30% PS (b and c), or PS (d) at a total concentration of 500 μg/ml. Samples were examined by negative stain TEM. Scale bar = 100 nm. C, as described in panel A, MxA was incubated in the absence or presence of liposomes generated from PC containing 10% or 30% PS, respectively. Liposomes generated from PC, PS, and TBL served as controls. Results from three independent experiments are shown.
FIGURE 7.
MxA loop L4 contains a putative membrane interacting motif. A, liposomes of negatively charged phospholipids protect L4 from proteolytic degradation. Aliquots of the MxA-liposome mixtures used in Fig. 6_A_ were subjected to proteinase K digestion as described in Fig. 5 and analyzed by Western blotting using anti-MxA- and anti-His tag-specific antibodies. B–D, liposome binding capacity of a stretch of positively charged amino acids in L4. MxA and the indicated MxA-L4 mutants (180 μg/ml) with single amino acid exchanges from lysine to glutamic acid were incubated in HCB300 in the absence (B) or presence of (C) GTPγS (100 μ
m
) or (D) TBL liposomes (1 mg/ml) for 20 min at 20 °C. After ultracentrifugation, the supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and Coomassie staining. Results of three independent experiments are shown. Error bars, S.D.
FIGURE 8.
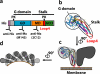
Structural model of MxA oligomers associated with liposomes. a, linear structure of human MxA with the N-terminal G domain and the C-terminal part forming the stalk. The recombinant protein is N-terminal fused to a His tag (His). Arrowheads indicate the positions of two proteinase K cleavage sites (PK). A red bar marks the position of loop L4 (position 533–572). The positions of the antibody binding sites for anti-His and anti-Mx antibodies, M143 and 2C12, are indicated by Y. b, hypothetical structure of a MxA monomer (side view) consisting of the MxA stalk fused to the G domain of dynamin. The red line indicates the position of loop L4. c, front view of an MxA oligomer associated with the membrane via loop L4 (red line), with the G domains sticking out in opposite directions leading to the typical T-bar structure. d, schematic side view of an MxA oligomeric ring structure associated with a lipid tubule (modified according to Ref. 8).
Similar articles
- Structural basis of oligomerization in the stalk region of dynamin-like MxA.
Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. Gao S, et al. Nature. 2010 May 27;465(7297):502-6. doi: 10.1038/nature08972. Epub 2010 Apr 28. Nature. 2010. PMID: 20428112 - Dynamin-like MxA GTPase: structural insights into oligomerization and implications for antiviral activity.
Haller O, Gao S, von der Malsburg A, Daumke O, Kochs G. Haller O, et al. J Biol Chem. 2010 Sep 10;285(37):28419-24. doi: 10.1074/jbc.R110.145839. Epub 2010 Jun 10. J Biol Chem. 2010. PMID: 20538602 Free PMC article. Review. - Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity.
Haller O, Kochs G. Haller O, et al. J Interferon Cytokine Res. 2011 Jan;31(1):79-87. doi: 10.1089/jir.2010.0076. Epub 2010 Dec 19. J Interferon Cytokine Res. 2011. PMID: 21166595 Review. - The dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes GTP to function in mitochondrial membrane fusion.
Meglei G, McQuibban GA. Meglei G, et al. Biochemistry. 2009 Mar 3;48(8):1774-84. doi: 10.1021/bi801723d. Biochemistry. 2009. PMID: 19236101
Cited by
- SMARCA2-regulated host cell factors are required for MxA restriction of influenza A viruses.
Dornfeld D, Dudek AH, Vausselin T, Günther SC, Hultquist JF, Giese S, Khokhlova-Cubberley D, Chew YC, Pache L, Krogan NJ, Garcia-Sastre A, Schwemmle M, Shaw ML. Dornfeld D, et al. Sci Rep. 2018 Feb 1;8(1):2092. doi: 10.1038/s41598-018-20458-2. Sci Rep. 2018. PMID: 29391557 Free PMC article. - A genetic model of differential susceptibility to human respiratory syncytial virus (RSV) infection.
Ciencewicki JM, Wang X, Marzec J, Serra ME, Bell DA, Polack FP, Kleeberger SR. Ciencewicki JM, et al. FASEB J. 2014 Apr;28(4):1947-56. doi: 10.1096/fj.13-239855. Epub 2014 Jan 13. FASEB J. 2014. PMID: 24421397 Free PMC article. - Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity.
Dick A, Graf L, Olal D, von der Malsburg A, Gao S, Kochs G, Daumke O. Dick A, et al. J Biol Chem. 2015 May 15;290(20):12779-92. doi: 10.1074/jbc.M115.650325. Epub 2015 Mar 31. J Biol Chem. 2015. PMID: 25829498 Free PMC article. - Membrane tethering and nucleotide-dependent conformational changes drive mitochondrial genome maintenance (Mgm1) protein-mediated membrane fusion.
Abutbul-Ionita I, Rujiviphat J, Nir I, McQuibban GA, Danino D. Abutbul-Ionita I, et al. J Biol Chem. 2012 Oct 26;287(44):36634-8. doi: 10.1074/jbc.C112.406769. Epub 2012 Sep 12. J Biol Chem. 2012. PMID: 22977249 Free PMC article. - Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein.
Fröhlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Fröhlich C, et al. EMBO J. 2013 May 2;32(9):1280-92. doi: 10.1038/emboj.2013.74. Epub 2013 Apr 12. EMBO J. 2013. PMID: 23584531 Free PMC article.
References
- Danino D., Hinshaw J. E. (2001) Current Opin. Cell Biol. 13, 454–460 - PubMed
- Praefcke G. J., McMahon H. T. (2004) Nat. Rev. Mol. Cell. Biol. 5, 133–147 - PubMed
- Haller O., Kochs G. (2011) J. Interferon Cytokine Res. 31, 79–87 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases