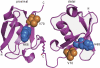Conformational dynamics of wild-type Lys-48-linked diubiquitin in solution - PubMed (original) (raw)
Conformational dynamics of wild-type Lys-48-linked diubiquitin in solution
Takashi Hirano et al. J Biol Chem. 2011.
Abstract
Proteasomal degradation is mediated through modification of target proteins by Lys-48-linked polyubiquitin (polyUb) chain, which interacts with several binding partners in this pathway through hydrophobic surfaces on individual Ub units. However, the previously reported crystal structures of Lys-48-linked diUb exhibit a closed conformation with sequestered hydrophobic surfaces. NMR studies on mutated Lys-48-linked diUb indicated a pH-dependent conformational equilibrium between closed and open states with the predominance of the former under neutral conditions (90% at pH 6.8). To address the question of how Ub-binding proteins can efficiently access the sequestered hydrophobic surfaces of Ub chains, we revisited the conformational dynamics of Lys-48-linked diUb in solution using wild-type diUb and cyclic forms of diUb in which the Ub units are connected through two Lys-48-mediated isopeptide bonds. Our newly determined crystal structure of wild-type diUb showed an open conformation, whereas NMR analyses of cyclic Lys-48-linked diUb in solution revealed that its structure resembled the closed conformation observed in previous crystal structures. Comparison of a chemical shift of wild-type diUb with that of monomeric Ub and cyclic diUb, which mimic the open and closed states, respectively, with regard to the exposure of hydrophobic surfaces to the solvent indicates that wild-type Lys-48-linked diUb in solution predominantly exhibits the open conformation (75% at pH 7.0), which becomes more populated upon lowering pH. The intrinsic properties of Lys-48-linked Ub chains to adopt the open conformation may be advantageous for interacting with Ub-binding proteins.
Figures
FIGURE 1.
The crystal structure of wild-type Lys-48-linked diUb. The proximal domain is located on the left, and the distal domain is located on the right. His-68 (blue) and Val-70 (orange) are shown in a space-filling representation. By superimposing the Cα carbons of the distal and proximal Ub units, an r.m.s.d. value of 0.2 Å was obtained for 73 residues, excluding the three C-terminal residues.
FIGURE 2.
1H-15N HSQC spectra of monomeric Ub (cyan), wild-type Lys-48-linked diUb (red), and cyclic Lys-48-linked diUb (black) at pH 7.0. The close views of spectral regions (boxed), including the peaks from Ile-13, Leu-43, Leu-69, and Val-70, are displayed at the bottom.
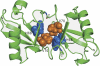
FIGURE 3.
Three-dimensional structural model of cyclic Lys-48-linked diUb derived from the NMR data. His-68 (blue) and Val-70 (orange) are shown in a space-filling representation.
FIGURE 4.
NMR characterization of conformational dynamics of Lys-48-linked diUb in solution. A, cartoon model of the conformational equilibrium of wild-type Lys-48-linked diUb. Cyclic Lys-48-linked diUb was used as a mimic of the closed state, whereas monomeric Ub was used as a mimic of the open state. The hydrophobic surface is colored red. The crystallographic data (
supplemental Fig. S2
) indicate that the distances between the two Ile-44 Cα atoms are 18.8 and 12.3 Å in the open and closed forms, respectively. B, 1H-15N HSQC peaks originating from Val-70 of monomeric Ub (cyan), wild-type Lys-48-linked diUb (red), and cyclic Lys-48-linked diUb (black) at the following different pH conditions: pH 7.0, 6.0, 5.0, and 4.6, starting from the left. C, pH dependence of the open conformation population of wild-type Lys-48-linked diUb (black) and its His-68→Val mutant (red) estimated on the basis of the dividing ratio of chemical shift differences of Val-70.
Similar articles
- Diubiquitin-Based NMR Analysis: Interactions Between Lys6-Linked diUb and UBA Domain of UBXN1.
Shahul Hameed D, van Tilburg GBA, Merkx R, Flierman D, Wienk H, El Oualid F, Hofmann K, Boelens R, Ovaa H. Shahul Hameed D, et al. Front Chem. 2020 Jan 22;7:921. doi: 10.3389/fchem.2019.00921. eCollection 2019. Front Chem. 2020. PMID: 32039147 Free PMC article. - NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains.
Hiranyakorn M, Yanaka S, Satoh T, Wilasri T, Jityuti B, Yagi-Utsumi M, Kato K. Hiranyakorn M, et al. Int J Mol Sci. 2020 Jul 28;21(15):5351. doi: 10.3390/ijms21155351. Int J Mol Sci. 2020. PMID: 32731397 Free PMC article. - Mutational and Environmental Effects on the Dynamic Conformational Distributions of Lys48-Linked Ubiquitin Chains.
Hiranyakorn M, Yagi-Utsumi M, Yanaka S, Ohtsuka N, Momiyama N, Satoh T, Kato K. Hiranyakorn M, et al. Int J Mol Sci. 2023 Mar 23;24(7):6075. doi: 10.3390/ijms24076075. Int J Mol Sci. 2023. PMID: 37047047 Free PMC article. - The lysine48-based polyubiquitin chain proteasomal signal: not a single child anymore.
Kravtsova-Ivantsiv Y, Sommer T, Ciechanover A. Kravtsova-Ivantsiv Y, et al. Angew Chem Int Ed Engl. 2013 Jan 2;52(1):192-8. doi: 10.1002/anie.201205656. Epub 2012 Nov 4. Angew Chem Int Ed Engl. 2013. PMID: 23124625 Review. - The Ball and Chain of Polyubiquitin Structures.
Alfano C, Faggiano S, Pastore A. Alfano C, et al. Trends Biochem Sci. 2016 Apr;41(4):371-385. doi: 10.1016/j.tibs.2016.01.006. Epub 2016 Feb 15. Trends Biochem Sci. 2016. PMID: 26899455 Review.
Cited by
- Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Gräwert TW, Knebel A, Svergun DI, Kulathu Y. Abdul Rehman SA, et al. Mol Cell. 2021 Oct 21;81(20):4176-4190.e6. doi: 10.1016/j.molcel.2021.08.024. Epub 2021 Sep 15. Mol Cell. 2021. PMID: 34529927 Free PMC article. - A snapshot of ubiquitin chain elongation: lysine 48-tetra-ubiquitin slows down ubiquitination.
Kovacev J, Wu K, Spratt DE, Chong RA, Lee C, Nayak J, Shaw GS, Pan ZQ. Kovacev J, et al. J Biol Chem. 2014 Mar 7;289(10):7068-7081. doi: 10.1074/jbc.M113.530576. Epub 2014 Jan 24. J Biol Chem. 2014. PMID: 24464578 Free PMC article. - Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch.
Dong X, Gong Z, Lu YB, Liu K, Qin LY, Ran ML, Zhang CL, Liu Z, Zhang WP, Tang C. Dong X, et al. Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):6770-6775. doi: 10.1073/pnas.1705718114. Epub 2017 Jun 13. Proc Natl Acad Sci U S A. 2017. PMID: 28611216 Free PMC article. - Diubiquitin-Based NMR Analysis: Interactions Between Lys6-Linked diUb and UBA Domain of UBXN1.
Shahul Hameed D, van Tilburg GBA, Merkx R, Flierman D, Wienk H, El Oualid F, Hofmann K, Boelens R, Ovaa H. Shahul Hameed D, et al. Front Chem. 2020 Jan 22;7:921. doi: 10.3389/fchem.2019.00921. eCollection 2019. Front Chem. 2020. PMID: 32039147 Free PMC article. - Promiscuous interactions of gp78 E3 ligase CUE domain with polyubiquitin chains.
Liu S, Chen Y, Li J, Huang T, Tarasov S, King A, Weissman AM, Byrd RA, Das R. Liu S, et al. Structure. 2012 Dec 5;20(12):2138-50. doi: 10.1016/j.str.2012.09.020. Epub 2012 Nov 1. Structure. 2012. PMID: 23123110 Free PMC article.
References
- Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 - PubMed
- Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 - PubMed
- Baboshina O. V., Haas A. L. (1996) J. Biol. Chem. 271, 2823–2831 - PubMed
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources