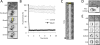The filament-forming protein Pil1 assembles linear eisosomes in fission yeast - PubMed (original) (raw)
The filament-forming protein Pil1 assembles linear eisosomes in fission yeast
Ruth Kabeche et al. Mol Biol Cell. 2011 Nov.
Abstract
The cortical cytoskeleton mediates a range of cellular activities such as endocytosis, cell motility, and the maintenance of cell rigidity. Traditional polymers, including actin, microtubules, and septins, contribute to the cortical cytoskeleton, but additional filament systems may also exist. In yeast cells, cortical structures called eisosomes generate specialized domains termed MCCs to cluster specific proteins at sites of membrane invaginations. Here we show that the core eisosome protein Pil1 forms linear cortical filaments in fission yeast cells and that purified Pil1 assembles into filaments in vitro. In cells, Pil1 cortical filaments are excluded from regions of cell growth and are independent of the actin and microtubule cytoskeletons. Pil1 filaments assemble slowly at the cell cortex and appear stable by time-lapse microscopy and fluorescence recovery after photobleaching. This stability does not require the cell wall, but Pil1 and the transmembrane protein Fhn1 colocalize and are interdependent for localization to cortical filaments. Increased Pil1 expression leads to cytoplasmic Pil1 rods that are stable and span the length of cylindrical fission yeast cells. We propose that Pil1 is a novel component of the yeast cytoskeleton, with implications for the role of filament assembly in the spatial organization of cells.
Figures
FIGURE 1:
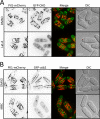
Pil1 forms filaments in fission yeast cells. (A) Western blot showing that Pil1-3HA is expressed in both mitotic and meiotic cells, whereas Pil2-3HA is expressed in meiotic cells but not mitotic cells. (B) Localization of Pil1-mCherry in vegetative cells. Left, inverted maximum projection for Z-planes in the top half of the cell. Asterisk, septating cell. Right, single focal planes and maximum projection for a single cell. (C) Localization of Pil1-mCherry and Pil2-mEGFP in mating cells. Top, inverted maximum projection from Z-planes in the top half of the cell; bottom, the middle focal plane. (D) Budding yeast Pil1 (_Sc_Pil1) forms filaments when expressed in fission yeast cells. Images show inverted single focal planes and maximum projection. (E) Fission yeast Pil1 (_Sp_Pil1) forms puncta when expressed in budding yeast cells. Images show inverted single focal planes and maximum projection. Scale bars: B–E, 5 μm.
FIGURE 2:
Pil1 filaments are independent of the actin and microtubule cytoskeletons. (A) Pil1 cortical filaments do not colocalize with or depend on the actin cytoskeleton. (B) Pil1 cortical filaments do not colocalize with or depend on microtubules. Images are maximum projections for Z-planes in the top half of the cell. Scale bars, 5 μm. Cells were treated with the indicated drug or DMSO control for 20 min before imaging.
FIGURE 3:
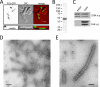
Pil1 forms filaments in bacterial cells and in vitro. (A) Localization of Pil1-GFP in E. coli cells. Fluorescence images are inverted maximum-intensity projections, and differential interference contrast (DIC) is red in the merged image. Scale bar, 1 μm. (B) Coomassie-stained SDS–polyacrylamide gel of purified Pil1, which migrates as a doublet similar to budding yeast Pil1 purified from bacteria. (C) Coomassie-stained SDS–polyacrylamide gel of purified Pil1 following centrifugation at high speed or low speed, as indicated. Pellet and supernatant fractions were separated and analyzed by SDS–PAGE. (D, E) Electron micrographs of purified Pil1 filaments. Samples were negatively stained and visualized by electron microscopy. Scale bar: D, 200 nm; E, 100 nm.
FIGURE 4:
Pil1 cortical filaments are static structures that assemble slowly. (A) Time-lapse microscopy of Pil1-mCherry during polarized cell growth. Images are inverted maximum projections for Z-planes in the top half of the cell. The green arrow highlights a newly assembled filament. (B) Time-lapse microscopy of Pil1-mCherry during cell division. The green arrow marks filament breakage. Scale bars, 5 μm. (C, D) Kymograph showing breakage and disassembly (C) or assembly (D) of single Pil1 filaments in cells. Images are inverted single focal planes from time-lapse microscopy.
FIGURE 5:
Pil1 cortical filaments are fixed structures that are independent of the cell wall. (A) FRAP of Pil1-mCherry. The yellow box denotes the bleached region, and images are inverted maximum projections for Z-planes in the top half of the cell. (B) Quantification of fluorescence recovery in bleached or control regions of FRAP experiments. Data points are the mean of 10 regions; error bars, SD. (C) Kymograph of single Pil1-mCherry filament bleached in the middle; each time point is 20 s. (D) Localization of Pil1-mCherry in emerging spheroplast. Image is inverted maximum projection; arrow, cell wall ghost. (E) Pil1-mCherry localizes to cortical filaments in spheroplasts. Images show inverted single focal planes and maximum projection, as indicated. Scale bars: A, D, E, 5 μm.
FIGURE 6:
Pil1 and Fhn1 are interdependent for localization. (A) Colocalization of Pil1-mCherry and Fhn1-mEGFP in cortical filaments; region boxed in white is magnified in the bottom row. (B) Localization of Fhn1-mEGFP in pil1Δ cells. (C) Localization of Pil1-mCherry in fhn1Δ cells. Note decreased abundance and length of cortical filaments. All images are inverted maximum projections from Z-planes in the top half of the cell; scale bars, 5 μm.
FIGURE 7:
Pil1 overexpression impairs cell polarity and cytokinesis. (A) Tenfold serial dilutions of cells containing the indicated plasmids. Expression was induced by growth on media lacking thiamine. (B) DIC images of cells containing control plasmid or Pil1 overexpression plasmid. Expression was induced by removal of thiamine from growth media for 20 h at 32°C. Scale bar, 10 μm. (C) Pil1 overexpression disrupts the localization of mitosis/cytokinesis protein Cdr2 and cell polarity proteins Pom1 and Tea1. The indicated cells were grown in the presence of thiamine (repressed) or in the absence of thiamine for 36 h at 25°C to induce Pil1 overexpression. Pil1 overexpression disrupts the concentration of Cdr2 in the cell middle and leads to loss of Pom1 and Tea1 from cell ends. Images are inverted maximum projections from Z-planes in the top half of cells. Scale bars, 5 μm.
FIGURE 8:
Formation of cytoplasmic rods by overexpressed Pil1. (A) Z-series and maximum projection of Pil1-GFP expressed by the medium-strength P41nmt1 promoter. (B) Pil1-GFP rods are stable in the absence of actin, microtubules, and septins. Cells were grown for 36 h at 32°C in the absence of thiamine and then treated with the indicated drug or DMSO control for 20 min. Images are maximum projections of Pil1-GFP overlaid on DIC image. Scale bars, 5 μm. (C) Time-lapse microscopy of cytoplasmic Pil1-GFP rods. Images are maximum projections; green arrow, mobile rod in cytoplasm. (D) FRAP analysis of cytoplasmic Pil1 rods. Kymograph shows time lapse for the rod outlined in gray. Yellow boxes in the prebleach image show sites of bleaching. Note the lack of movement for unbleached region in the center of the rod.
Similar articles
- Seg1 controls eisosome assembly and shape.
Moreira KE, Schuck S, Schrul B, Fröhlich F, Moseley JB, Walther TC, Walter P. Moreira KE, et al. J Cell Biol. 2012 Aug 6;198(3):405-20. doi: 10.1083/jcb.201202097. J Cell Biol. 2012. PMID: 22869600 Free PMC article. - A Pil1-Sle1-Syj1-Tax4 functional pathway links eisosomes with PI(4,5)P2 regulation.
Kabeche R, Roguev A, Krogan NJ, Moseley JB. Kabeche R, et al. J Cell Sci. 2014 Mar 15;127(Pt 6):1318-26. doi: 10.1242/jcs.143545. Epub 2014 Jan 16. J Cell Sci. 2014. PMID: 24434583 Free PMC article. - Assembly of fission yeast eisosomes in the plasma membrane of budding yeast: import of foreign membrane microdomains.
Vaskovicova K, Stradalova V, Efenberk A, Opekarova M, Malinsky J. Vaskovicova K, et al. Eur J Cell Biol. 2015 Jan;94(1):1-11. doi: 10.1016/j.ejcb.2014.10.003. Epub 2014 Oct 22. Eur J Cell Biol. 2015. PMID: 25457676 - Fission yeast cytoskeletons and cell polarity factors: connecting at the cortex.
La Carbona S, Le Goff C, Le Goff X. La Carbona S, et al. Biol Cell. 2006 Nov;98(11):619-31. doi: 10.1042/BC20060048. Biol Cell. 2006. PMID: 17042740 Review. - The yeast actin cytoskeleton.
Mishra M, Huang J, Balasubramanian MK. Mishra M, et al. FEMS Microbiol Rev. 2014 Mar;38(2):213-27. doi: 10.1111/1574-6976.12064. Epub 2014 Feb 20. FEMS Microbiol Rev. 2014. PMID: 24467403 Review.
Cited by
- Effects of FSGS-associated mutations on the stability and function of myosin-1 in fission yeast.
Bi J, Carroll RT, James ML, Ouderkirk JL, Krendel M, Sirotkin V. Bi J, et al. Dis Model Mech. 2015 Aug 1;8(8):891-902. doi: 10.1242/dmm.020214. Epub 2015 Jun 18. Dis Model Mech. 2015. PMID: 26092123 Free PMC article. - Plasma Membrane MCC/Eisosome Domains Promote Stress Resistance in Fungi.
Lanze CE, Gandra RM, Foderaro JE, Swenson KA, Douglas LM, Konopka JB. Lanze CE, et al. Microbiol Mol Biol Rev. 2020 Sep 16;84(4):e00063-19. doi: 10.1128/MMBR.00063-19. Print 2020 Nov 18. Microbiol Mol Biol Rev. 2020. PMID: 32938742 Free PMC article. Review. - Pil1 cytoplasmic rods contain bundles of crosslinked tubules.
Kabeche R, Howard L, Moseley JB. Kabeche R, et al. Commun Integr Biol. 2015 Mar 4;8(1):e990848. doi: 10.4161/19420889.2014.990848. eCollection 2015 Jan-Feb. Commun Integr Biol. 2015. PMID: 26609339 Free PMC article. - Plasma Membrane Protein Nce102 Modulates Morphology and Function of the Yeast Vacuole.
Vaskovicova K, Vesela P, Zahumensky J, Folkova D, Balazova M, Malinsky J. Vaskovicova K, et al. Biomolecules. 2020 Oct 23;10(11):1476. doi: 10.3390/biom10111476. Biomolecules. 2020. PMID: 33114062 Free PMC article. - Megadalton-node assembly by binding of Skb1 to the membrane anchor Slf1.
Deng L, Kabeche R, Wang N, Wu JQ, Moseley JB. Deng L, et al. Mol Biol Cell. 2014 Sep 1;25(17):2660-8. doi: 10.1091/mbc.E14-04-0896. Epub 2014 Jul 9. Mol Biol Cell. 2014. PMID: 25009287 Free PMC article.
References
- Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, Paoletti A. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr Biol. 2009;19:961–966. - PubMed
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
- Brach T, Specht T, Kaksonen M. Reassessment of the role of plasma membrane domains in the regulation of vesicular traffic in yeast. J Cell Sci. 2011;124:328–337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases