Using network analysis to explore co-occurrence patterns in soil microbial communities - PubMed (original) (raw)
Using network analysis to explore co-occurrence patterns in soil microbial communities
Albert Barberán et al. ISME J. 2012 Feb.
Erratum in
- ISME J. 2014 Apr;8(4):952
Abstract
Exploring large environmental datasets generated by high-throughput DNA sequencing technologies requires new analytical approaches to move beyond the basic inventory descriptions of the composition and diversity of natural microbial communities. In order to investigate potential interactions between microbial taxa, network analysis of significant taxon co-occurrence patterns may help to decipher the structure of complex microbial communities across spatial or temporal gradients. Here, we calculated associations between microbial taxa and applied network analysis approaches to a 16S rRNA gene barcoded pyrosequencing dataset containing >160 000 bacterial and archaeal sequences from 151 soil samples from a broad range of ecosystem types. We described the topology of the resulting network and defined operational taxonomic unit categories based on abundance and occupancy (that is, habitat generalists and habitat specialists). Co-occurrence patterns were readily revealed, including general non-random association, common life history strategies at broad taxonomic levels and unexpected relationships between community members. Overall, we demonstrated the potential of exploring inter-taxa correlations to gain a more integrated understanding of microbial community structure and the ecological rules guiding community assembly.
Figures
Figure 1
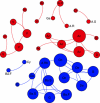
Network of co-occurring 90% cutoff OTUs based on correlation analysis. A connection stands for a strong (Spearman's _ρ_>0.6) and significant (_P_-value <0.01) correlation. The size of each node is proportional to the number of connections (that is, degree). Left panel: OTUs colored by taxonomy. Right panel: OTUs colored by abundance and occupancy (generalists and specialists).
Figure 2
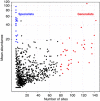
Abundance (y axis) and occupancy (x axis) plot for the 90% cutoff OTUs. Habitat generalists OTUs (in red) defined as appearing in >80 soil samples. Habitat specialists OTUs (in blue) defined as locally abundant (>18 sequences) and appearance in <10 soils.
Figure 3
Network of co-occurring generalists and specialists 90% cutoff OTUs based on correlation analysis. A connection stands for a strong (Spearman's _ρ_>0.6) and significant (_P_-value <0.01) correlation. The size of each node is proportional to the number of connections (that is, degree). Labels according to taxonomic affiliation: Ac, Acidobacteria. A.R, Alphaproteobacteria; Rhizobiales. A.Rh, Alphaproteobacteria; Rhodobacterales. A.S, Alphaproteobacteria; Sphingomonadales. Ba.F, Bacteroidetes; Flavobacteria. Ba.S, Bacteroidetes; Sphingobacteria. Ch, Chloroflexi. Cr, Crenarchaeota. Cy, Cyanobacteria; D, Deltaproteobacteria. De, Deinococcus. G, Gemmatimonadetes. Ga, Gammaproteobacteria. V, Verrucomicrobia.
Figure 4
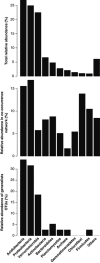
Relative abundance of different microbial taxonomic groups. Top panel: number of sequences in all soil samples. Middle panel: number of significant co-occurrent OTUs (nodes from Figure 2). Low panel: number of cosmopolitan OTUs.
Similar articles
- Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2.
Zhou J, Deng Y, Luo F, He Z, Yang Y. Zhou J, et al. mBio. 2011 Jul 26;2(4):e00122-11. doi: 10.1128/mBio.00122-11. Print 2011. mBio. 2011. PMID: 21791581 Free PMC article. - Contrasting spatial patterns and ecological attributes of soil bacterial and archaeal taxa across a landscape.
Constancias F, Saby NP, Terrat S, Dequiedt S, Horrigue W, Nowak V, Guillemin JP, Biju-Duval L, Chemidlin Prévost-Bouré N, Ranjard L. Constancias F, et al. Microbiologyopen. 2015 Jun;4(3):518-31. doi: 10.1002/mbo3.256. Epub 2015 Apr 28. Microbiologyopen. 2015. PMID: 25922908 Free PMC article. - Altitudinal distribution patterns of soil bacterial and archaeal communities along mt. Shegyla on the Tibetan Plateau.
Wang JT, Cao P, Hu HW, Li J, Han LL, Zhang LM, Zheng YM, He JZ. Wang JT, et al. Microb Ecol. 2015 Jan;69(1):135-45. doi: 10.1007/s00248-014-0465-7. Epub 2014 Jul 30. Microb Ecol. 2015. PMID: 25074792 - Combined phylogenetic and genomic approaches for the high-throughput study of microbial habitat adaptation.
Zaneveld JR, Parfrey LW, Van Treuren W, Lozupone C, Clemente JC, Knights D, Stombaugh J, Kuczynski J, Knight R. Zaneveld JR, et al. Trends Microbiol. 2011 Oct;19(10):472-82. doi: 10.1016/j.tim.2011.07.006. Epub 2011 Aug 25. Trends Microbiol. 2011. PMID: 21872475 Free PMC article. Review. - From diversity to complexity: Microbial networks in soils.
Guseva K, Darcy S, Simon E, Alteio LV, Montesinos-Navarro A, Kaiser C. Guseva K, et al. Soil Biol Biochem. 2022 Jun;169:108604. doi: 10.1016/j.soilbio.2022.108604. Soil Biol Biochem. 2022. PMID: 35712047 Free PMC article. Review.
Cited by
- Determining indicator taxa across spatial and seasonal gradients in the Columbia River coastal margin.
Fortunato CS, Eiler A, Herfort L, Needoba JA, Peterson TD, Crump BC. Fortunato CS, et al. ISME J. 2013 Oct;7(10):1899-911. doi: 10.1038/ismej.2013.79. Epub 2013 May 30. ISME J. 2013. PMID: 23719153 Free PMC article. - High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats.
Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. Zhou J, et al. mBio. 2015 Jan 27;6(1):e02288-14. doi: 10.1128/mBio.02288-14. mBio. 2015. PMID: 25626903 Free PMC article. Review. - Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes.
Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM, Zhang T. Li B, et al. ISME J. 2015 Nov;9(11):2490-502. doi: 10.1038/ismej.2015.59. Epub 2015 Apr 28. ISME J. 2015. PMID: 25918831 Free PMC article. - Roles of Thermophiles and Fungi in Bitumen Degradation in Mostly Cold Oil Sands Outcrops.
Wong ML, An D, Caffrey SM, Soh J, Dong X, Sensen CW, Oldenburg TB, Larter SR, Voordouw G. Wong ML, et al. Appl Environ Microbiol. 2015 Oct;81(19):6825-38. doi: 10.1128/AEM.02221-15. Epub 2015 Jul 24. Appl Environ Microbiol. 2015. PMID: 26209669 Free PMC article. - Effects of the invasion of Ralstonia solanacearum on soil microbial community structure in Wuhan, China.
Wu Q-Y, Ma R, Wang X, Ma Y-N, Wang Z-S, Wei H-L, Zhang X-X. Wu Q-Y, et al. mSphere. 2024 Feb 28;9(2):e0066523. doi: 10.1128/msphere.00665-23. Epub 2024 Jan 17. mSphere. 2024. PMID: 38231250 Free PMC article.
References
- Auguet JC, Barberán A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. - PubMed
- Barberán A, Casamayor EO. Global phylogenetic community structure and beta-diversity patterns of surface bacterioplankton metacommunities. Aquat Microb Ecol. 2010;59:1–10.
- Barberán A, Casamayor EO. Euxinic freshwater hypolimnia promote bacterial endemicity in continental areas. Microb Ecol. 2011;61:465–472. - PubMed
- Bastian M, Heymann S, Jacomy M. Gephi: An Open Source Software For Exploring and Manipulating Networks. In International AAAI conference on weblogs and social media: San Jose, California; 2009.