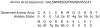Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone - PubMed (original) (raw)
Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone
Jason Trigg et al. PLoS One. 2011.
Abstract
The alpha-helical coiled coil can adopt a variety of topologies, among the most common of which are parallel and antiparallel dimers and trimers. We present Multicoil2, an algorithm that predicts both the location and oligomerization state (two versus three helices) of coiled coils in protein sequences. Multicoil2 combines the pairwise correlations of the previous Multicoil method with the flexibility of Hidden Markov Models (HMMs) in a Markov Random Field (MRF). The resulting algorithm integrates sequence features, including pairwise interactions, through multinomial logistic regression to devise an optimized scoring function for distinguishing dimer, trimer and non-coiled-coil oligomerization states; this scoring function is used to produce Markov Random Field potentials that incorporate pairwise correlations localized in sequence. Multicoil2 significantly improves both coiled-coil detection and dimer versus trimer state prediction over the original Multicoil algorithm retrained on a newly-constructed database of coiled-coil sequences. The new database, comprised of 2,105 sequences containing 124,088 residues, includes reliable structural annotations based on experimental data in the literature. Notably, the enhanced performance of Multicoil2 is evident when tested in stringent leave-family-out cross-validation on the new database, reflecting expected performance on challenging new prediction targets that have minimal sequence similarity to known coiled-coil families. The Multicoil2 program and training database are available for download from http://multicoil2.csail.mit.edu.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
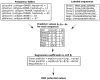
Figure 1. Flow-chart overview of the Multicoil2 method.
From the training set of labelled dimer, trimer and negative coiled coil sequences, we compute the probability of each amino acid at each heptad position in dimer sequences and trimer sequences. Also, we compute the probability of each amino acid in negative sequences. From the resulting frequency tables shown in the upper left, along with the training sequences shown in the upper right, we compute sequence features for each training sequence. Running a multinomial logistic regression on these values generates a three-way classifier which is used in the MRF as described in Methods.
Figure 2. Dimer versus trimer recognition.
Multicoil2 and Multicoil ROC curves based on leave-family-out cross validation for per-residue (a) and per-sequence (b) recognition.
Figure 3. Coiled coil detection.
Multicoil2, retrained Multicoil and Paircoil2 ROC curves based on leave-family-out cross validation for per-residue (a) and per-sequence (b) detection. Note the very small values on x-axis for false positive rate.
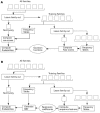
Figure 4. Overview of training of the Multicoil and Multicoil2 algorithms.
(a) The raw scores used to generate the Multicoil gaussians for each of n-1 families are computed based on frequency tables generated from the other n-2 families. (b) The Multicoil2 sequence features for each of n-1 families are computed based on frequency tables generated from the other n-2 families. Those features are used to find the regression coefficients, which determine the MRF.
Figure 5. Graph of allowed transitions.
There are two copies of each state, one for the dimer state and one for the trimer state (the transitions of the trimer states are omitted - they are identical to the dimer transitions). The “0” nodes at the top and bottom of the figure refer to the same non-coiled-coil state.
Figure 6. Example of hidden states corresponding to a given amino acid sequence in the Multicoil2 model.
Similar articles
- MultiCoil: a program for predicting two- and three-stranded coiled coils.
Wolf E, Kim PS, Berger B. Wolf E, et al. Protein Sci. 1997 Jun;6(6):1179-89. doi: 10.1002/pro.5560060606. Protein Sci. 1997. PMID: 9194178 Free PMC article. - Computational characterization of parallel dimeric and trimeric coiled-coils using effective amino acid indices.
Li C, Wang XF, Chen Z, Zhang Z, Song J. Li C, et al. Mol Biosyst. 2015 Feb;11(2):354-60. doi: 10.1039/c4mb00569d. Epub 2014 Dec 1. Mol Biosyst. 2015. PMID: 25435395 - SCORER 2.0: an algorithm for distinguishing parallel dimeric and trimeric coiled-coil sequences.
Armstrong CT, Vincent TL, Green PJ, Woolfson DN. Armstrong CT, et al. Bioinformatics. 2011 Jul 15;27(14):1908-14. doi: 10.1093/bioinformatics/btr299. Epub 2011 May 16. Bioinformatics. 2011. PMID: 21576179 - Critical evaluation of in silico methods for prediction of coiled-coil domains in proteins.
Li C, Ching Han Chang C, Nagel J, Porebski BT, Hayashida M, Akutsu T, Song J, Buckle AM. Li C, et al. Brief Bioinform. 2016 Mar;17(2):270-82. doi: 10.1093/bib/bbv047. Epub 2015 Jul 15. Brief Bioinform. 2016. PMID: 26177815 Free PMC article. Review. - The design of coiled-coil structures and assemblies.
Woolfson DN. Woolfson DN. Adv Protein Chem. 2005;70:79-112. doi: 10.1016/S0065-3233(05)70004-8. Adv Protein Chem. 2005. PMID: 15837514 Review.
Cited by
- Novel nuclear targeting coiled-coil protein of Helicobacter pylori showing Ca(2+)-independent, Mg(2+)-dependent DNase I activity.
Kwon YC, Kim S, Lee YS, Lee JC, Cho MJ, Lee WK, Kang HL, Song JY, Baik SC, Ro HS. Kwon YC, et al. J Microbiol. 2016 May;54(5):387-95. doi: 10.1007/s12275-016-5631-9. Epub 2016 Apr 20. J Microbiol. 2016. PMID: 27095458 - PKD-dependent PARP12-catalyzed mono-ADP-ribosylation of Golgin-97 is required for E-cadherin transport from Golgi to plasma membrane.
Grimaldi G, Filograna A, Schembri L, Lo Monte M, Di Martino R, Pirozzi M, Spano D, Beccari AR, Parashuraman S, Luini A, Valente C, Corda D. Grimaldi G, et al. Proc Natl Acad Sci U S A. 2022 Jan 4;119(1):e2026494119. doi: 10.1073/pnas.2026494119. Proc Natl Acad Sci U S A. 2022. PMID: 34969853 Free PMC article. - Cross-linking mass spectrometry identifies new interfaces of Augmin required to localise the γ-tubulin ring complex to the mitotic spindle.
Chen JWC, Chen ZA, Rogala KB, Metz J, Deane CM, Rappsilber J, Wakefield JG. Chen JWC, et al. Biol Open. 2017 May 15;6(5):654-663. doi: 10.1242/bio.022905. Biol Open. 2017. PMID: 28351835 Free PMC article. - The Structure and Topology of α-Helical Coiled Coils.
Lupas AN, Bassler J, Dunin-Horkawicz S. Lupas AN, et al. Subcell Biochem. 2017;82:95-129. doi: 10.1007/978-3-319-49674-0_4. Subcell Biochem. 2017. PMID: 28101860 Free PMC article. Review. - Evolution of three parent genes and their retrogene copies in Drosophila species.
O'Neill RS, Clark DV. O'Neill RS, et al. Int J Evol Biol. 2013;2013:693085. doi: 10.1155/2013/693085. Epub 2013 Jun 5. Int J Evol Biol. 2013. PMID: 23841016 Free PMC article.
References
- Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163:258–269. - PubMed
- Craig AW, Zirngibl R, Greer P. Disruption of coiled-coil domains in Fer protein-tyrosine kinase abolishes trimerization but not kinase activation. J Biol Chem. 1999;274:19934–19942. - PubMed
- Gruber M, Soding J, Lupas AN. Comparative analysis of coiled-coil prediction methods. J Struct Biol. 2006;155:140–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous