The first identification of lysine malonylation substrates and its regulatory enzyme - PubMed (original) (raw)
. 2011 Dec;10(12):M111.012658.
doi: 10.1074/mcp.M111.012658. Epub 2011 Sep 9.
Zhike Lu, Zhongyu Xie, Zhongyi Cheng, Yue Chen, Minjia Tan, Hao Luo, Yi Zhang, Wendy He, Ke Yang, Bernadette M M Zwaans, Daniel Tishkoff, Linh Ho, David Lombard, Tong-Chuan He, Junbiao Dai, Eric Verdin, Yang Ye, Yingming Zhao
Affiliations
- PMID: 21908771
- PMCID: PMC3237090
- DOI: 10.1074/mcp.M111.012658
The first identification of lysine malonylation substrates and its regulatory enzyme
Chao Peng et al. Mol Cell Proteomics. 2011 Dec.
Abstract
Protein post-translational modifications (PTMs) at the lysine residue, such as lysine methylation, acetylation, and ubiquitination, are diverse, abundant, and dynamic. They play a key role in the regulation of diverse cellular physiology. Here we report discovery of a new type of lysine PTM, lysine malonylation (Kmal). Kmal was initially detected by mass spectrometry and protein sequence-database searching. The modification was comprehensively validated by Western blot, tandem MS, and high-performance liquid chromatography of synthetic peptides, isotopic labeling, and identification of multiple Kmal substrate proteins. Kmal is a dynamic and evolutionarily conserved PTM observed in mammalian cells and bacterial cells. In addition, we demonstrate that Sirt5, a member of the class III lysine deacetylases, can catalyze lysine demalonylation and lysine desuccinylation reactions both in vitro and in vivo. This result suggests the possibility of nondeacetylation activity of other class III lysine deacetylases, especially those without obvious acetylation protein substrates. Our results therefore reveal a new type of PTM pathway and identify the first enzyme that can regulate lysine malonylation and lysine succinylation status.
Figures
Fig. 1.
Chemical structures of lysine, acetyllysine, succinyllisine, malonyllysine, and the candidate CoA for lysine malonylation.
Fig. 2.
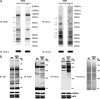
Detection of lysine malonylation by Western blot. A, Specificity of the anti-Kmal antibody. Dot-blot assay was carried out using anti-Kmal antibody by incubation with peptide libraries bearing a fixed unmodified K, Kac, Kbuty, Ksucc, Kmal, two glycine-rich peptides (CEGGGGKmalGGSG) bearing a unmodified K (GGK), and a malonylated K (GGKmal), respectively. Each peptide library contains 13 residues CXXXXXKXXXXXX, where X is a mixture of 19 amino acids (excluding cysteine), C is cysteine, and the seventh residue is a fixed lysine residue: unmodified lysine (K), acetyllysine (Kac), butyryllysine (Kbuty), succinyllysine (Ksucc), malonyllysine (Kmal). GGK indicates a glycine-rich peptide, CEGGGGKGGSG, whereas GGKmal is a malonylated glycine-rich peptide, CEGGGGKmalGGSG, in which the lysine residue is malonylated. B, Western blot of the whole cell lysates from HeLa cell and E. coli. Western blot was performed using the whole cell lysates from HeLa cell and E. coli with competition of modified CEGGGGKmalGGSG, unmodified CEGGGGKGGSG, succinylated BSA, or peptide library bearing a succinylated lysine(Ksucc).
Fig. 3.
Verification of Kmal peptides by HPLC/MS/MS of synthetic peptides. A, High-resolution CID MS/MS spectra of an α-enolase tryptic peptide (TAIGK+85.9983Da AGYTDK) with a mass shift of + 85.9983 Da (top), the synthetic TAIGKmalAGYTDK (middle), and their mixture (bottom). Inset shows the mass-to-charge ratios (m/z) of the precursor ions. A neutral loss of CO2 (44 Da) indicated decarboxylation. B, Extracted ion chromatograms (XICs) of the _in vivo_-derived peptide (top), its synthetic counterpart (middle), and their mixture (bottom). C, High-resolution CID MS/MS spectra of a PGK1 tryptic peptide (FHVEEEGK+85.9985DaGK) with a mass shift of + 85.9985 Da (top), the synthetic FHVEEEGKmalGK (middle), and their mixture (bottom). Inset shows m/z of precursor ions. D, XICs of the _in vivo_-derived peptide (top), its synthetic counterpart (middle) and their mixture (bottom).
Fig. 4.
Isotopic labeling of Kmal peptides. A, Western blot of whole cell lysates from HeLa cells treated with (+) or without (-) malonate. Western blot was performed with competition of either modified CEGGGGKmalGGSG or unmodified CEGGGGKGGSG. B, CID MS/MS spectra of a 12C-Kmal peptide, LAADVGK12C-malAGAER (top). The neutral loss of the carbon dioxide (CO2) is designated as “-44”; MS/MS spectra (bottom) of a K13C-mal peptide, LAADVGK13C-malAGAER, from HeLa cells treated with [1, 2, 3-13C] malonate. The neutral loss of the carbon-13 dioxide (13CO2) is designated as “–45”.
Fig. 5.
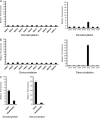
Sirt5 is an in vitro enzyme of lysine demalonylation and lysine desuccinylation. Fluorometric assay to detect in vitro lysine demalonylation activities (A) and lysine desuccinylation activities (B) among HDACs. C, Fluorometric assay to compare demalonylation and desuccinylation activities between wt Sirt5 and its inactive mutant.
Fig. 6.
Lysine demalonylation and desuccinylation activities of Sirt5 in peptide substrates. A, LC/MS detection of in vitro demalonylated peptide. A synthetic peptide, AFNQGKmalIFK (HRMS, m/z, 569.7982), from TUT1 was used as the substrate. B, LC/MS detection of an in vitro desuccinylated peptide. A synthetic peptide, VLLPEYGGTKsuccVVLDDK (HRMS, m/z 923.4968), from HSPE1 was used as substrate. The assays were carried out without Sirt5, with wt Sirt5, with Sirt5 inactive mutant (H158Y), without NAD+, with Sirt5 in the presence of nicotinamide (40 m
m
), sirtinol (200 μ
m
), or TSA (2 μ
m
). A triangle and a diamond indicate modified and unmodified peptides, respectively.
Fig. 7.
Sirt5 can catalyze protein lysine demalonylation and desuccinylation reactions both in vitro and in vivo. A, Sirt5 can catalyze in vitro demalonylation and desuccinylation reactions in mitochondrial proteins. Thirty micrograms of HeLa mitochondrial proteins were resolved by SDS-PAGE gel and transferred to the PVDF membranes. The membranes were incubated with or without 80 μg of Sirt5 in 2 ml of SDAC buffer (25 m
m
Tris·Cl, 130 m
m
NaCl, 3.0 m
m
KCl, 1 m
m
MgCl2, 1 m
m
DTT, 0.1% PEG8000, pH = 8.0) containing 0.5 m
m
NAD+ at 37 °C overnight. Then the membranes were blocked and probed with anti-Kmal antibody and anti-Ksucc antibody, respectively. Western blot for SOD-2 was used as a loading control. The arrows indicate the protein bands whose lysine malonylation or lysine succinylation are changed in response to Sirt5. B, Mice lacking SIRT5 show increased global lysine malonylation and succinylation, but no change in protein acetylation. Livers from a Sirt5 −/− (KO) mouse and its wild type littermate control were homogenized and examined for global lysine malonylation, succinylation, and acetylation by Western blot. Ponceau salt staining as well as Western blot for α-tubulin and ETF (mitochondrial electron transfer flavoprotein) were used as loading controls.
Similar articles
- Lysine succinylation and lysine malonylation in histones.
Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Xie Z, et al. Mol Cell Proteomics. 2012 May;11(5):100-7. doi: 10.1074/mcp.M111.015875. Epub 2012 Mar 4. Mol Cell Proteomics. 2012. PMID: 22389435 Free PMC article. - Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli.
Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang Y, Wan X, Chen Y, Cha YH, Lin H, Zhao Y, Tan M. Colak G, et al. Mol Cell Proteomics. 2013 Dec;12(12):3509-20. doi: 10.1074/mcp.M113.031567. Epub 2013 Oct 31. Mol Cell Proteomics. 2013. PMID: 24176774 Free PMC article. - Systematic analysis of lysine malonylation in Streptococcus mutans.
Li Z, Wu Q, Zhang Y, Zhou X, Peng X. Li Z, et al. Front Cell Infect Microbiol. 2022 Nov 28;12:1078572. doi: 10.3389/fcimb.2022.1078572. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36519128 Free PMC article. - Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation.
Hirschey MD, Zhao Y. Hirschey MD, et al. Mol Cell Proteomics. 2015 Sep;14(9):2308-15. doi: 10.1074/mcp.R114.046664. Epub 2015 Feb 25. Mol Cell Proteomics. 2015. PMID: 25717114 Free PMC article. Review. - Understanding the Function of Mammalian Sirtuins and Protein Lysine Acylation.
Wang M, Lin H. Wang M, et al. Annu Rev Biochem. 2021 Jun 20;90:245-285. doi: 10.1146/annurev-biochem-082520-125411. Epub 2021 Apr 13. Annu Rev Biochem. 2021. PMID: 33848425 Review.
Cited by
- CHHM: a Manually Curated Catalogue of Human Histone Modifications Revealing Hotspot Regions and Unique Distribution Patterns.
Ma W, Ding X, Xu J, Poon TCW. Ma W, et al. Int J Biol Sci. 2024 Jul 2;20(10):3760-3772. doi: 10.7150/ijbs.95954. eCollection 2024. Int J Biol Sci. 2024. PMID: 39113691 Free PMC article. - SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase.
Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, Haigis MC. Laurent G, et al. Mol Cell. 2013 Jun 6;50(5):686-98. doi: 10.1016/j.molcel.2013.05.012. Mol Cell. 2013. PMID: 23746352 Free PMC article. - SIRT3 Activation a Promise in Drug Development? New Insights into SIRT3 Biology and Its Implications on the Drug Discovery Process.
Lambona C, Zwergel C, Valente S, Mai A. Lambona C, et al. J Med Chem. 2024 Feb 8;67(3):1662-1689. doi: 10.1021/acs.jmedchem.3c01979. Epub 2024 Jan 23. J Med Chem. 2024. PMID: 38261767 Free PMC article. Review. - System-wide studies of N-lysine acetylation in Rhodopseudomonas palustris reveal substrate specificity of protein acetyltransferases.
Crosby HA, Pelletier DA, Hurst GB, Escalante-Semerena JC. Crosby HA, et al. J Biol Chem. 2012 May 4;287(19):15590-601. doi: 10.1074/jbc.M112.352104. Epub 2012 Mar 13. J Biol Chem. 2012. PMID: 22416131 Free PMC article. - Lipid-induced mitochondrial stress and insulin action in muscle.
Muoio DM, Neufer PD. Muoio DM, et al. Cell Metab. 2012 May 2;15(5):595-605. doi: 10.1016/j.cmet.2012.04.010. Cell Metab. 2012. PMID: 22560212 Free PMC article.
References
- Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 - PubMed
- Walsh C. T., Garneau-Tsodikova S., Gatto G. J., Jr. (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. Int. Ed Engl. 44, 7342–7372 - PubMed
- Johnson S. A., Hunter T. (2005) Kinomics: methods for deciphering the kinome. Nat. Methods 2, 17–25 - PubMed
- Denu J. M., Dixon J. E. (1998) Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 2, 633–641 - PubMed
- Guarente L. (2007) Sirtuins in aging and disease. Cold Spring Harb. Symp Quant Biol. 72, 483–488 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous