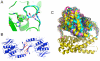RosettaRemodel: a generalized framework for flexible backbone protein design - PubMed (original) (raw)
RosettaRemodel: a generalized framework for flexible backbone protein design
Po-Ssu Huang et al. PLoS One. 2011.
Abstract
We describe RosettaRemodel, a generalized framework for flexible protein design that provides a versatile and convenient interface to the Rosetta modeling suite. RosettaRemodel employs a unified interface, called a blueprint, which allows detailed control over many aspects of flexible backbone protein design calculations. RosettaRemodel allows the construction and elaboration of customized protocols for a wide range of design problems ranging from loop insertion and deletion, disulfide engineering, domain assembly, loop remodeling, motif grafting, symmetrical units, to de novo structure modeling.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist. YAB currently works at Arzeda Corporation, but declares no competing interest. YAB's involvement with this project was during the time he was a postdoc at the University of Washington. His use of the method described in the manuscript will have to follow the general licensing scheme like all other commercial users.
Figures
Figure 1. Examples of backbone manipulation using RosettaRemodel.
In the center is the crystal structure of protein G (PDB ID: 1PGA), which was used as the starting point for all the different cases. The colored regions highlights changed made with RosettaRemodel.
Figure 2. Blueprint Assignment Examples.
Figure 3. Constraints file and its associated blueprint file.
Each block in the constraints file is represented in the blueprint file with the CST1A, CST1B, CST2A and CST2B notation. The enzdes constraint format is discussed in Richter et al. in detail. In this figure we show the association between cst and blueprint files. Each enzdes constraint block, defined between CST::BEGIN and CST::END statements, always contains two elements, and to interface with blueprint, the first definition is defined as “A” and the second as “B,” and each element requires a corresponding assignment in the blueprint. Each block is also associated with a numerical value from 1 to the total number of blocks defined in the cst file. In this example, the first constraint pair (CST1A/CST1B) is used to restraint one of the residues (residue 17) on the strand being built (residues 16–20) to within a hydrogen bonding distance with a stationary residue (residue 12). The second constraint pair (CST2A/CST2B) operates on the sidechains of residue 9 and 20 for hydrogen bonding between the functional groups. The distinction between a backbone and sidechain definition is the choice of atom types using Rosetta atom type names and a required “is_backbone” statement because enzdes constraint protocol does not automatically treat atoms as backbone by names. In this example, the hydrogen bonding constraint is defined for a pair of atoms within 2.8+/−0.2 Å, with a force constant of 100. The trailing “0” in the distanceAB definition is for non-covalent interaction.
Figure 4. Examples of designs made with RosettaRemodel.
A) A cysteine protease site, with cys-his intermediate shown in green sticks. The loop in cyan was rebuilt from its original position (in green) to introduce sidechain-directed stabilization of the oxyanion in the intermediate. The re-designed model uses an asparagine in direct contact with the oxyanion instead of the wild-type aspartic acid (in pink). B) Two interacting loops in a symmetry arrangement were rebuilt to increase interactions across the subunits. C) Domain localization. A domain assembly designed with a pair of linkers. In this figure, the ensemble of an internally inserted domain is shown moving relative to the stationary structure that hosts the insertion as a result of sampling the loops linking them. With this type of sampling, one could model the localization of the final assembly.
Similar articles
- A computational method for the design of nested proteins by loop-directed domain insertion.
Blacklock KM, Yang L, Mulligan VK, Khare SD. Blacklock KM, et al. Proteins. 2018 Mar;86(3):354-369. doi: 10.1002/prot.25445. Epub 2018 Jan 24. Proteins. 2018. PMID: 29250820 - Integration of the Rosetta suite with the python software stack via reproducible packaging and core programming interfaces for distributed simulation.
Ford AS, Weitzner BD, Bahl CD. Ford AS, et al. Protein Sci. 2020 Jan;29(1):43-51. doi: 10.1002/pro.3721. Epub 2019 Dec 2. Protein Sci. 2020. PMID: 31495995 Free PMC article. - Predicting the tolerated sequences for proteins and protein interfaces using RosettaBackrub flexible backbone design.
Smith CA, Kortemme T. Smith CA, et al. PLoS One. 2011;6(7):e20451. doi: 10.1371/journal.pone.0020451. Epub 2011 Jul 18. PLoS One. 2011. PMID: 21789164 Free PMC article. - Designing protein structures and complexes with the molecular modeling program Rosetta.
Kuhlman B. Kuhlman B. J Biol Chem. 2019 Dec 13;294(50):19436-19443. doi: 10.1074/jbc.AW119.008144. Epub 2019 Nov 7. J Biol Chem. 2019. PMID: 31699898 Free PMC article. Review. - Protocols for Molecular Modeling with Rosetta3 and RosettaScripts.
Bender BJ, Cisneros A 3rd, Duran AM, Finn JA, Fu D, Lokits AD, Mueller BK, Sangha AK, Sauer MF, Sevy AM, Sliwoski G, Sheehan JH, DiMaio F, Meiler J, Moretti R. Bender BJ, et al. Biochemistry. 2016 Aug 30;55(34):4748-63. doi: 10.1021/acs.biochem.6b00444. Epub 2016 Aug 16. Biochemistry. 2016. PMID: 27490953 Free PMC article. Review.
Cited by
- Structures and dynamics of the novel S1/S2 protease cleavage site loop of the SARS-CoV-2 spike glycoprotein.
Lemmin T, Kalbermatter D, Harder D, Plattet P, Fotiadis D. Lemmin T, et al. J Struct Biol X. 2020;4:100038. doi: 10.1016/j.yjsbx.2020.100038. Epub 2020 Oct 5. J Struct Biol X. 2020. PMID: 33043289 Free PMC article. - The coming of age of de novo protein design.
Huang PS, Boyken SE, Baker D. Huang PS, et al. Nature. 2016 Sep 15;537(7620):320-7. doi: 10.1038/nature19946. Nature. 2016. PMID: 27629638 Review. - Computational design of a Diels-Alderase from a thermophilic esterase: the importance of dynamics.
Linder M, Johansson AJ, Olsson TS, Liebeschuetz J, Brinck T. Linder M, et al. J Comput Aided Mol Des. 2012 Sep;26(9):1079-95. doi: 10.1007/s10822-012-9601-y. Epub 2012 Sep 16. J Comput Aided Mol Des. 2012. PMID: 22983490 - Computational strategies for the design of new enzymatic functions.
Świderek K, Tuñón I, Moliner V, Bertran J. Świderek K, et al. Arch Biochem Biophys. 2015 Sep 15;582:68-79. doi: 10.1016/j.abb.2015.03.013. Epub 2015 Mar 19. Arch Biochem Biophys. 2015. PMID: 25797438 Free PMC article. Review. - Dissecting the stability determinants of a challenging de novo protein fold using massively parallel design and experimentation.
Kim TE, Tsuboyama K, Houliston S, Martell CM, Phoumyvong CM, Lemak A, Haddox HK, Arrowsmith CH, Rocklin GJ. Kim TE, et al. Proc Natl Acad Sci U S A. 2022 Oct 11;119(41):e2122676119. doi: 10.1073/pnas.2122676119. Epub 2022 Oct 3. Proc Natl Acad Sci U S A. 2022. PMID: 36191185 Free PMC article.
References
- Cochran FV, Wu SP, Wang W, Nanda V, Saven JG, et al. Computational de novo design and characterization of a four-helix bundle protein that selectively binds a nonbiological cofactor. Journal of the American Chemical Society. 2005;127:1346–1347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources