SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics - PubMed (original) (raw)
SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics
Filip Bielejec et al. Bioinformatics. 2011.
Abstract
Summary: SPREAD is a user-friendly, cross-platform application to analyze and visualize Bayesian phylogeographic reconstructions incorporating spatial-temporal diffusion. The software maps phylogenies annotated with both discrete and continuous spatial information and can export high-dimensional posterior summaries to keyhole markup language (KML) for animation of the spatial diffusion through time in virtual globe software. In addition, SPREAD implements Bayes factor calculation to evaluate the support for hypotheses of historical diffusion among pairs of discrete locations based on Bayesian stochastic search variable selection estimates. SPREAD takes advantage of multicore architectures to process large joint posterior distributions of phylogenies and their spatial diffusion and produces visualizations as compelling and interpretable statistical summaries for the different spatial projections.
Availability: SPREAD is licensed under the GNU Lesser GPL and its source code is freely available as a GitHub repository: https://github.com/phylogeography/SPREAD CONTACT: filip.bielejec@rega.kuleuven.be.
Figures
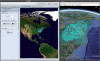
Fig. 1.
Screenshot of SPREAD. This example visualizes the phylogeographic history of rabies among raccoons along the Eastern US seaboard under a continuous diffusion model. Such visualizations allow users to quickly inspect key evolutionary changes in their geographic context. Further generation of KML output enables interactive exploration in the time dimension as well in freely available virtual globe software, such as
Google Earth
(on the right).
Similar articles
- spread.gl: visualizing pathogen dispersal in a high-performance browser application.
Li Y, Bollen N, Hong SL, Brusselmans M, Gambaro F, Klaps J, Suchard MA, Rambaut A, Lemey P, Dellicour S, Baele G. Li Y, et al. Bioinformatics. 2024 Nov 28;40(12):btae721. doi: 10.1093/bioinformatics/btae721. Bioinformatics. 2024. PMID: 39626311 Free PMC article. - Bayesian phylogeography finds its roots.
Lemey P, Rambaut A, Drummond AJ, Suchard MA. Lemey P, et al. PLoS Comput Biol. 2009 Sep;5(9):e1000520. doi: 10.1371/journal.pcbi.1000520. Epub 2009 Sep 25. PLoS Comput Biol. 2009. PMID: 19779555 Free PMC article. - SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes.
Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. Bielejec F, et al. Mol Biol Evol. 2016 Aug;33(8):2167-9. doi: 10.1093/molbev/msw082. Epub 2016 Apr 23. Mol Biol Evol. 2016. PMID: 27189542 Free PMC article. - Phylowood: interactive web-based animations of biogeographic and phylogeographic histories.
Landis MJ, Bedford T. Landis MJ, et al. Bioinformatics. 2014 Jan 1;30(1):123-4. doi: 10.1093/bioinformatics/btt635. Epub 2013 Nov 4. Bioinformatics. 2014. PMID: 24191071 Free PMC article. - Contrasting the epidemiological and evolutionary dynamics of influenza spatial transmission.
Viboud C, Nelson MI, Tan Y, Holmes EC. Viboud C, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Feb 4;368(1614):20120199. doi: 10.1098/rstb.2012.0199. Print 2013 Mar 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23382422 Free PMC article. Review.
Cited by
- Phylodynamics of H5N1 Highly Pathogenic Avian Influenza in Europe, 2005-2010: Potential for Molecular Surveillance of New Outbreaks.
Alkhamis MA, Moore BR, Perez AM. Alkhamis MA, et al. Viruses. 2015 Jun 23;7(6):3310-28. doi: 10.3390/v7062773. Viruses. 2015. PMID: 26110587 Free PMC article. - Genotype-specific variation in West Nile virus dispersal in California.
Duggal NK, Reisen WK, Fang Y, Newman RM, Yang X, Ebel GD, Brault AC. Duggal NK, et al. Virology. 2015 Nov;485:79-85. doi: 10.1016/j.virol.2015.07.004. Epub 2015 Jul 25. Virology. 2015. PMID: 26210076 Free PMC article. - Visualizing time-related data in biology, a review.
Secrier M, Schneider R. Secrier M, et al. Brief Bioinform. 2014 Sep;15(5):771-82. doi: 10.1093/bib/bbt021. Epub 2013 Apr 12. Brief Bioinform. 2014. PMID: 23585583 Free PMC article. Review. - Phylodynamics and Coat Protein Analysis of Babaco Mosaic Virus in Ecuador.
Mosquera-Yuqui F, Flores FJ, Moncayo EA, Garzón-Proaño BA, Méndez MA, Guevara FE, Quito-Avila DF, Viera W, Cornejo-Franco JF, Izquierdo AR, Noceda C. Mosquera-Yuqui F, et al. Plants (Basel). 2022 Jun 22;11(13):1646. doi: 10.3390/plants11131646. Plants (Basel). 2022. PMID: 35807598 Free PMC article. - Combination of surveillance tools reveals that Yellow Fever virus can remain in the same Atlantic Forest area at least for three transmission seasons.
Abreu FVS, Delatorre E, Dos Santos AAC, Ferreira-de-Brito A, de Castro MG, Ribeiro IP, Furtado ND, Vargas WP, Ribeiro MS, Meneguete P, Bonaldo MC, Bello G, Lourenço-de-Oliveira R. Abreu FVS, et al. Mem Inst Oswaldo Cruz. 2019;114:e190076. doi: 10.1590/0074-02760190076. Epub 2019 Apr 29. Mem Inst Oswaldo Cruz. 2019. PMID: 31038550 Free PMC article.
References
- Kidd D.M., Ritchie M.G. Phylogeographic information systems: putting the geography into phylogeography. J. Biogeogr. 2006;33:1851–1865.
Publication types
MeSH terms
Grants and funding
- 095831/WT_/Wellcome Trust/United Kingdom
- 260864/ERC_/European Research Council/International
- R01 GM086887/GM/NIGMS NIH HHS/United States
- R01 HG006139/HG/NHGRI NIH HHS/United States