Imaging proteins inside cells with fluorescent tags - PubMed (original) (raw)
Review
Imaging proteins inside cells with fluorescent tags
Georgeta Crivat et al. Trends Biotechnol. 2012 Jan.
Abstract
Watching biological molecules provides clues to their function and regulation. Some of the most powerful methods of labeling proteins for imaging use genetically encoded fluorescent fusion tags. There are four standard genetic methods of covalently tagging a protein with a fluorescent probe for cellular imaging. These use (i) autofluorescent proteins, (ii) self-labeling enzymes, (iii) enzymes that catalyze the attachment of a probe to a target sequence, and (iv) biarsenical dyes that target tetracysteine motifs. Each of these techniques has advantages and disadvantages. In this review, we cover new developments in these methods and discuss practical considerations for their use in imaging proteins inside living cells.
Published by Elsevier Ltd.
Figures
Figure 1
Auto-fluorescent proteins as fusion tags. a) Table of excitation and emission wavelengths for twelve optimized fluorescent proteins (FP). The comparative brightness of each FP is listed as the product of the quantum yield and the extinction coefficient at the peak absorbance wavelength divided by 1000. Values were generating from the literature [1, 23, 26, 28]. b) Structure of EGFP [39]. A magnified view of the cyclized chromophore (TYG) is shown to the right. c) Cartoon of N- and C- terminal linkers introduced during genetic fusion of a protein of interest (POI) to an FP in standard Clontech vectors. The minimum and maximum linkers are indicated by parenthesis. The unstructured amino acids from EGFP are colored green. A common flexible peptide linker is shown below.
Figure 2
Self-labeling enzymes as fluorescent fusion tags. a) The structure of AGT/SNAP tag (top) and the Haloalkane dehydrogenase tag (bottom). The residues that are labeled by the fluorescent ligand are shown in yellow. In Halo, the mutated catalytic histidine is shown in orange. b) (top) Structure of a benzyl guanine-linked TMR ligand for SNAP lag labeling and a (bottom) TMR-linked haloalkane for Halotag labeling along with c) the corresponding reaction mechanisms for covalent attachment.
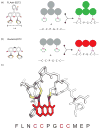
Figure 3
Structure of the biarsenical dyes FLAsH-EDT2 (a) and ReAsH-EDT2 (b). (c) Model of the optimized tetracysteine peptide bound to ReAsH based on the NMR structure of the complex [70].
Figure 4
Examples of fluorescent labeling methods in cells. a) Total Internal Reflection Fluorescence image (TIRF) of a living PC12 cells expressing the F-actin binding protein ITPKA tagged with tdTomato [85]. b) TIRF image of a PC12 cell expressing Halotag-Beta actin labeled with the red fluorophore TMR-halotag ligand. c) Confocal image of an erythrocyte infected with transgenic Plasmodium falciparum expressing the tetracysteine tag (TC)-containing protein KAHRP (+His)-TC labeled with ReAsH.
Similar articles
- Chemical tags: applications in live cell fluorescence imaging.
Wombacher R, Cornish VW. Wombacher R, et al. J Biophotonics. 2011 Jun;4(6):391-402. doi: 10.1002/jbio.201100018. Epub 2011 May 12. J Biophotonics. 2011. PMID: 21567974 Review. - Fluorescent labelling in living cells.
Schneider AFL, Hackenberger CPR. Schneider AFL, et al. Curr Opin Biotechnol. 2017 Dec;48:61-68. doi: 10.1016/j.copbio.2017.03.012. Epub 2017 Apr 7. Curr Opin Biotechnol. 2017. PMID: 28395178 Review. - [Visualization and Functional Regulation of Live Cell Proteins Based on Labeling Probe Design].
Mizukami S, Kikuchi K. Mizukami S, et al. Yakugaku Zasshi. 2016;136(1):21-7. doi: 10.1248/yakushi.15-00225-4. Yakugaku Zasshi. 2016. PMID: 26725663 Review. Japanese. - Multicolor protein labeling in living cells using mutant β-lactamase-tag technology.
Watanabe S, Mizukami S, Hori Y, Kikuchi K. Watanabe S, et al. Bioconjug Chem. 2010 Dec 15;21(12):2320-6. doi: 10.1021/bc100333k. Epub 2010 Oct 20. Bioconjug Chem. 2010. PMID: 20961132 - Genetic tags for labelling live cells: gap junctions and beyond.
Falk M. Falk M. Trends Cell Biol. 2002 Sep;12(9):399-404. doi: 10.1016/s0962-8924(02)02343-7. Trends Cell Biol. 2002. PMID: 12220850
Cited by
- Engineered Proteins and Materials Utilizing Residue-Specific Noncanonical Amino Acid Incorporation.
Majekodunmi T, Britton D, Montclare JK. Majekodunmi T, et al. Chem Rev. 2024 Aug 14;124(15):9113-9135. doi: 10.1021/acs.chemrev.3c00855. Epub 2024 Jul 15. Chem Rev. 2024. PMID: 39008623 Free PMC article. Review. - A vector system for single and tandem expression of cloned genes and multi-colour fluorescent tagging in Haloferax volcanii.
Ithurbide S, de Silva RT, Brown HJ, Shinde V, Duggin IG. Ithurbide S, et al. Microbiology (Reading). 2024 May;170(5):001461. doi: 10.1099/mic.0.001461. Microbiology (Reading). 2024. PMID: 38787390 Free PMC article. - Recent Developments of Hybrid Fluorescence Techniques: Advances in Amyloid Detection Methods.
Prasanna AM, Sen P. Prasanna AM, et al. Curr Protein Pept Sci. 2024;25(9):667-681. doi: 10.2174/0113892037291597240429094515. Curr Protein Pept Sci. 2024. PMID: 38715332 Review. - Degron tagging for rapid protein degradation in mice.
Hernández-Morán BA, Taylor G, Lorente-Macías Á, Wood AJ. Hernández-Morán BA, et al. Dis Model Mech. 2024 Apr 1;17(4):dmm050613. doi: 10.1242/dmm.050613. Epub 2024 Apr 26. Dis Model Mech. 2024. PMID: 38666498 Free PMC article. - Cell-Type-Dependent Recruitment Dynamics of FUS Protein at Laser-Induced DNA Damage Sites.
Niu Y, Pal A, Szewczyk B, Japtok J, Naumann M, Glaß H, Hermann A. Niu Y, et al. Int J Mol Sci. 2024 Mar 20;25(6):3526. doi: 10.3390/ijms25063526. Int J Mol Sci. 2024. PMID: 38542501 Free PMC article.
References
- Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. - PubMed
- Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. - PubMed
- Juillerat A, et al. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol. 2003;10:313–317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources