Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates - PubMed (original) (raw)
Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates
Antoine Molaro et al. Cell. 2011.
Abstract
During germ cell and preimplantation development, mammalian cells undergo nearly complete reprogramming of DNA methylation patterns. We profiled the methylomes of human and chimp sperm as a basis for comparison to methylation patterns of ESCs. Although the majority of promoters escape methylation in both ESCs and sperm, the corresponding hypomethylated regions show substantial structural differences. Repeat elements are heavily methylated in both germ and somatic cells; however, retrotransposons from several subfamilies evade methylation more effectively during male germ cell development, whereas other subfamilies show the opposite trend. Comparing methylomes of human and chimp sperm revealed a subset of differentially methylated promoters and strikingly divergent methylation in retrotransposon subfamilies, with an evolutionary impact that is apparent in the underlying genomic sequence. Thus, the features that determine DNA methylation patterns differ between male germ cells and somatic cells, and elements of these features have diverged between humans and chimpanzees.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
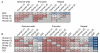
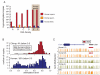
Figure 1. A global view of sperm and ESC methylomes
A. Correlations between methylomes with methylation levels measured at individual CpG sites. Correlations are displayed for CpGs genome wide, within promoters and within repeats, and correlation coefficients are colored blue to red to indicate low to high, respectively. B. Overlap between sets of HMRs from human sperm, chimp sperm and ESC methylomes, along with annotated CGIs. Each cell gives the fraction of HMRs corresponding to the row that overlap HMRs corresponding to the column. Colors are overlaid as in A. See also Supplementary Tables 1
Figure 2. Differentially reprogrammed genes and their functions
A. Average methylation through promoters (−1kbp to +1kbp) in human sperm and ESCs based on RefSeq gene annotations. Promoters that were hypomethylated only in sperm are shown in blue, only in ESCs in red, and promoters methylated in both are shaded orange. B. Average methylation of promoters associated with GO terms found enriched in the sperm specific hypomethylated fraction (see A), with the addition of genes from the “embryonic development” term. Individual genes involved in the “piRNA metabolic process” are indicated as an example. See also Supplementary Figure 1 and Supplementary Tables 2
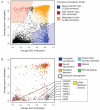
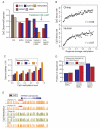
Figure 3. Characteristics of HMRs emerging from germline and somatic reprogramming
A. Log-scale plot depicting the sizes (in bases) and number of CpGs for all identified HMRs in human sperm (left), chimp sperm (middle) and human ESCs (right). Diagonal lines indicate 10% CpG density (in green) and 1% CpG density (dashed line). HMRs are colored according to promoter overlap (red), overlap with repeats but not promoters (blue), or overlap with neither (orange). B. Average methylation around all TSS overlapping HMRs in both sperm (orange) and ESC (blue); solid lines represent data smoothed using a 20 base sliding window. A schematic depicts the concepts of extended and nested HMRs at promoters. C. Average methylation at the −5 to +5 CpGs around boundaries of extended sperm HMRs and nested ESC HMRs (with the +1 CpG defined as the first inside an HMR on either side). D. Ratios of observed-to-expected CpG density for each nucleotide position relative to boundaries of extended sperm HMRs (left) and nested ESC HMRs (right). Solid lines indicate values smoothed using a 20 base sliding window. E. Average inter-CpG distance for −5 to +5 CpGs around HMR boundaries of extended sperm and nested ESC HMRs. F. Ratio of observed to expected frequencies of the CACGTG pattern at −5 to +5 CpGs for extended sperm and nested ESC HMRs. See also Supplementary Figure 2 and Supplementary Table 3
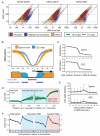
Figure 4. Differential repeat methylation during male germ cell and somatic reprogramming
A. For each repeat class, the proportion of elements that overlap HMRs is shown for human sperm (red), chimp sperm (orange) and ESCs (blue). B. Upper: Sliding 10kb window average methylation level across chromosome 12 (red) and satellite density (blue). Lower: Chromosome 12 centromeric region with HMRs (blue) and methylation level (orange) for human sperm and ESCs. C. CpG densities of hypomethylated repeat copies (red) and methylated repeat copies (yellow) for LINEs, LTRs, SINEs and SVAs. D. HMR overlap distribution around full length L1PA2 and LTR12 ERV9 elements for human sperm (blue) and ESCs (red). See also Supplementary Figure 3 and Supplementary Tables 4
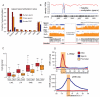
Figure 5. Divergent methylation of SVA elements between human and chimp
A. Proportion of hypomethylated SVA copies hypomethylated according to subfamily (A to F) for human sperm (red), chimp sperm (orange) and ESCs (blue). B. The distribution of average methylation levels is shown for 358 human (lower) and chimp (upper) SVAs forming high-confidence orthologous pairs. C. An SVA insertion shared by human and chimp but with differential methylation between species.
Figure 6. Sequence features associated with methylome divergence
A. Ratio of observed-to-expected CpG density across all HMRs, those overlapping promoters, those sperm- or ESC-specific, and the extended/nested HMRs. Data for sperm is indicated in blue and for ESC is indicated in red; orange indicates ratio immediately outside extended HMRs. B. Frequency of regions under CpG decay as a function of methylation for both human and chimp at locations of HMRs in the other species. Decay is presented for chimp in the upper panel and for human in the lower panel. C. Proportion of sequences displaying over 1% nucleotide divergence relative to the inferred ancestor using gorilla as an out-group and counting only non -CpG sites. D. The promoter of the human HTR3E (serotonin receptor) gene contains an HMR in both human donors but in neither chimp donor. See also Supplementary Figure 5 and Supplementary Tables 5
Similar articles
- Evolution of the sperm methylome of primates is associated with retrotransposon insertions and genome instability.
Fukuda K, Inoguchi Y, Ichiyanagi K, Ichiyanagi T, Go Y, Nagano M, Yanagawa Y, Takaesu N, Ohkawa Y, Imai H, Sasaki H. Fukuda K, et al. Hum Mol Genet. 2017 Sep 15;26(18):3508-3519. doi: 10.1093/hmg/ddx236. Hum Mol Genet. 2017. PMID: 28637190 - Evolutionary expansion of DNA hypomethylation in the mammalian germline genome.
Qu J, Hodges E, Molaro A, Gagneux P, Dean MD, Hannon GJ, Smith AD. Qu J, et al. Genome Res. 2018 Feb;28(2):145-158. doi: 10.1101/gr.225896.117. Epub 2017 Dec 19. Genome Res. 2018. PMID: 29259021 Free PMC article. - Comparative whole genome DNA methylation profiling of cattle sperm and somatic tissues reveals striking hypomethylated patterns in sperm.
Zhou Y, Connor EE, Bickhart DM, Li C, Baldwin RL, Schroeder SG, Rosen BD, Yang L, Van Tassell CP, Liu GE. Zhou Y, et al. Gigascience. 2018 May 1;7(5):giy039. doi: 10.1093/gigascience/giy039. Gigascience. 2018. PMID: 29635292 Free PMC article. - Germline-derived DNA methylation and early embryo epigenetic reprogramming: The selected survival of imprints.
Monk D. Monk D. Int J Biochem Cell Biol. 2015 Oct;67:128-38. doi: 10.1016/j.biocel.2015.04.014. Epub 2015 May 9. Int J Biochem Cell Biol. 2015. PMID: 25966912 Review. - DNA methylation in epigenetic inheritance of metabolic diseases through the male germ line.
Illum LRH, Bak ST, Lund S, Nielsen AL. Illum LRH, et al. J Mol Endocrinol. 2018 Feb;60(2):R39-R56. doi: 10.1530/JME-17-0189. Epub 2017 Dec 4. J Mol Endocrinol. 2018. PMID: 29203518 Review.
Cited by
- An epigenetic memory of pregnancy in the mouse mammary gland.
Dos Santos CO, Dolzhenko E, Hodges E, Smith AD, Hannon GJ. Dos Santos CO, et al. Cell Rep. 2015 May 19;11(7):1102-9. doi: 10.1016/j.celrep.2015.04.015. Epub 2015 May 7. Cell Rep. 2015. PMID: 25959817 Free PMC article. - Mammalian non-CG methylations are conserved and cell-type specific and may have been involved in the evolution of transposon elements.
Guo W, Zhang MQ, Wu H. Guo W, et al. Sci Rep. 2016 Aug 30;6:32207. doi: 10.1038/srep32207. Sci Rep. 2016. PMID: 27573482 Free PMC article. - Evolutionary consequences of DNA methylation on the GC content in vertebrate genomes.
Mugal CF, Arndt PF, Holm L, Ellegren H. Mugal CF, et al. G3 (Bethesda). 2015 Jan 15;5(3):441-7. doi: 10.1534/g3.114.015545. G3 (Bethesda). 2015. PMID: 25591920 Free PMC article. - New insights into establishment and maintenance of DNA methylation imprints in mammals.
Kelsey G, Feil R. Kelsey G, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20110336. doi: 10.1098/rstb.2011.0336. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166397 Free PMC article. Review. - Shape-based alignment of genomic landscapes in multi-scale resolution.
Ashida H, Asai K, Hamada M. Ashida H, et al. Nucleic Acids Res. 2012 Aug;40(14):6435-48. doi: 10.1093/nar/gks354. Epub 2012 May 4. Nucleic Acids Res. 2012. PMID: 22561376 Free PMC article.
References
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. - PubMed
- Bantysh OB, Buzdin AA. Novel family of human transposable elements formed due to fusion of the first exon of gene MAST2 with retrotransposon SVA. Biochemistry (Mosc) 2009;74:1393–1399. - PubMed
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. - PubMed
Publication types
MeSH terms
Grants and funding
- RC2 HD064459-02/HD/NICHD NIH HHS/United States
- RC2 HD064459/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01HG005238/HG/NHGRI NIH HHS/United States
- RC2 HD064459-01/HD/NICHD NIH HHS/United States
- R01 HG005238/HG/NHGRI NIH HHS/United States
- 1RC2HD064459/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources