Bambus 2: scaffolding metagenomes - PubMed (original) (raw)
Bambus 2: scaffolding metagenomes
Sergey Koren et al. Bioinformatics. 2011.
Abstract
Motivation: Sequencing projects increasingly target samples from non-clonal sources. In particular, metagenomics has enabled scientists to begin to characterize the structure of microbial communities. The software tools developed for assembling and analyzing sequencing data for clonal organisms are, however, unable to adequately process data derived from non-clonal sources.
Results: We present a new scaffolder, Bambus 2, to address some of the challenges encountered when analyzing metagenomes. Our approach relies on a combination of a novel method for detecting genomic repeats and algorithms that analyze assembly graphs to identify biologically meaningful genomic variants. We compare our software to current assemblers using simulated and real data. We demonstrate that the repeat detection algorithms have higher sensitivity than current approaches without sacrificing specificity. In metagenomic datasets, the scaffolder avoids false joins between distantly related organisms while obtaining long-range contiguity. Bambus 2 represents a first step toward automated metagenomic assembly.
Availability: Bambus 2 is open source and available from http://amos.sf.net.
Contact: mpop@umiacs.umd.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
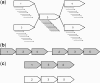
Fig. 1.
(a) The unitig graph representation of a single unitig, 3, having double the coverage of the surrounding unitigs. Solid black arrows correspond to reads comprising a unitig. (b) One of the possible resolutions of the graph presented in (a). This example places unitig 3 in two locations along a single genome. (c) A second of the possible resolutions of the graph presented in (a). This example places unitig 3 at the same location in two genomes (highlighted in different colors).
Fig. 2.
(a) A variant motif detected on the Sim3 dataset. The motif corresponds to a ferrochelatase gene in E.coli. There are two alternate versions of the gene within the E.coli K12 (2338) and E.coli O157:H7 (2034) genomes. (b) A CLUSTAL W (Thompson et al., 1994) alignment of a subset of the fasta output from Bambus 2, with an edit region corresponding to (a).
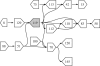
Fig. 3.
The figure shows a subset of a bacterial assembly where nodes are connected if they share paired-end reads. The shaded node, 119, is a repeat that occurs on many shortest paths.
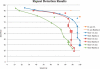
Fig. 4.
Repeat detection comparison. Ideal repeat detection corresponds to the top-right corner of the graph, with 100% sensitivity and specificity. We vary the genome size estimate (a critical parameter in the procedure for detecting repeats) for CA, generating a curve for each dataset. The CA-met default is indicated by large shaded points. The Bambus 2 repeat detection is fully automated, generating a single point. As CA is designed for clonal organisms, only the default genome size estimate is used for B.suis. The gold standard is built from REPuter. All tests are run using the set of unitigs generated by CA-met. Sensitivity:  . Specificity:
. Specificity:  .
.
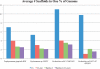
Fig. 5.
Assembly results for three simulated datasets. The _y_-axis represents the minimum number of scaffolds that add up to 1% of the genome size. Lower bars represent a better assembly. Bambus 2 produces large scaffolds for a wide range of coverage levels in our simulated datasets. Bambus 2 (CA-met) is Bambus 2 run using CA-met instead of using Minimus unitigs. We aligned the assembly (all contigs >2 kb) to the reference and counted coverage by reciprocal best matches over 95% identity. We use reciprocal best matches to avoid double counting Bambus 2 motifs that cover the same genomic region. We divide the number of scaffolds by the genome coverage and average the results, by genome, on all three simulated datasets to evaluate performance across varying coverage.
Fig. 6.
Assembly results for the acid mine metagenomic dataset. The _y_-axis represents the minimum number of scaffolds that add up to 1% of the genome size. Lower bars represent a better assembly. Bambus 2 produces larger scaffolds that CA-met in three of the five genomes. We calculated assembly statistics as in Figure 5. In three genomes, both CA and Bambus 2 produced slightly >100% coverage. This is due to redundancy within the MUMmer alignments.
Similar articles
- Exploring variation-aware contig graphs for (comparative) metagenomics using MaryGold.
Nijkamp JF, Pop M, Reinders MJ, de Ridder D. Nijkamp JF, et al. Bioinformatics. 2013 Nov 15;29(22):2826-34. doi: 10.1093/bioinformatics/btt502. Epub 2013 Sep 20. Bioinformatics. 2013. PMID: 24058058 Free PMC article. - GraphBin: refined binning of metagenomic contigs using assembly graphs.
Mallawaarachchi V, Wickramarachchi A, Lin Y. Mallawaarachchi V, et al. Bioinformatics. 2020 Jun 1;36(11):3307-3313. doi: 10.1093/bioinformatics/btaa180. Bioinformatics. 2020. PMID: 32167528 - Metagenomic assembly through the lens of validation: recent advances in assessing and improving the quality of genomes assembled from metagenomes.
Olson ND, Treangen TJ, Hill CM, Cepeda-Espinoza V, Ghurye J, Koren S, Pop M. Olson ND, et al. Brief Bioinform. 2019 Jul 19;20(4):1140-1150. doi: 10.1093/bib/bbx098. Brief Bioinform. 2019. PMID: 28968737 Free PMC article. - Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.
Wang Z, Wang Y, Fuhrman JA, Sun F, Zhu S. Wang Z, et al. Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review. - Web Resources for Metagenomics Studies.
Dudhagara P, Bhavsar S, Bhagat C, Ghelani A, Bhatt S, Patel R. Dudhagara P, et al. Genomics Proteomics Bioinformatics. 2015 Oct;13(5):296-303. doi: 10.1016/j.gpb.2015.10.003. Epub 2015 Nov 18. Genomics Proteomics Bioinformatics. 2015. PMID: 26602607 Free PMC article. Review.
Cited by
- Sequence assembly demystified.
Nagarajan N, Pop M. Nagarajan N, et al. Nat Rev Genet. 2013 Mar;14(3):157-67. doi: 10.1038/nrg3367. Epub 2013 Jan 29. Nat Rev Genet. 2013. PMID: 23358380 Review. - InteMAP: Integrated metagenomic assembly pipeline for NGS short reads.
Lai B, Wang F, Wang X, Duan L, Zhu H. Lai B, et al. BMC Bioinformatics. 2015 Aug 7;16:244. doi: 10.1186/s12859-015-0686-x. BMC Bioinformatics. 2015. PMID: 26250558 Free PMC article. - Approaches for in silico finishing of microbial genome sequences.
Kremer FS, McBride AJA, Pinto LS. Kremer FS, et al. Genet Mol Biol. 2017 Jul-Sep 01;40(3):553-576. doi: 10.1590/1678-4685-GMB-2016-0230. Genet Mol Biol. 2017. PMID: 28898352 Free PMC article. - HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads.
Nurk S, Walenz BP, Rhie A, Vollger MR, Logsdon GA, Grothe R, Miga KH, Eichler EE, Phillippy AM, Koren S. Nurk S, et al. Genome Res. 2020 Sep;30(9):1291-1305. doi: 10.1101/gr.263566.120. Epub 2020 Aug 14. Genome Res. 2020. PMID: 32801147 Free PMC article. - Graph-based self-supervised learning for repeat detection in metagenomic assembly.
Azizpour A, Balaji A, Treangen TJ, Segarra S. Azizpour A, et al. Genome Res. 2024 Oct 11;34(9):1468-1476. doi: 10.1101/gr.279136.124. Genome Res. 2024. PMID: 39029947
References
- Altschul S, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed