adegenet 1.3-1: new tools for the analysis of genome-wide SNP data - PubMed (original) (raw)
adegenet 1.3-1: new tools for the analysis of genome-wide SNP data
Thibaut Jombart et al. Bioinformatics. 2011.
Abstract
Summary: While the R software is becoming a standard for the analysis of genetic data, classical population genetics tools are being challenged by the increasing availability of genomic sequences. Dedicated tools are needed for harnessing the large amount of information generated by next-generation sequencing technologies. We introduce new tools implemented in the adegenet 1.3-1 package for handling and analyzing genome-wide single nucleotide polymorphism (SNP) data. Using a bit-level coding scheme for SNP data and parallelized computation, adegenet enables the analysis of large genome-wide SNPs datasets using standard personal computers.
Availability: adegenet 1.3-1 is available from CRAN: http://cran.r-project.org/web/packages/adegenet/. Information and support including a dedicated forum of discussion can be found on the adegenet website: http://adegenet.r-forge.r-project.org/. adegenet is released with a manual and four tutorials totalling over 300 pages of documentation, and distributed under the GNU General Public Licence (≥2).
Contact: t.jombart@imperial.ac.uk.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
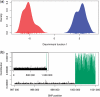
Fig. 1.
DAPC of simulated data (see text). (a) Density of individual scores on the first discriminant function, with groups represented in red and blue. (b) SNP contribution to the separation of the groups; the last (structured) 1000 SNPs are coloured in green; the last 4000 SNPs are represented in the main plot, while the figure corresponding to all SNPs is shown in inset.
Similar articles
- adegenet: a R package for the multivariate analysis of genetic markers.
Jombart T. Jombart T. Bioinformatics. 2008 Jun 1;24(11):1403-5. doi: 10.1093/bioinformatics/btn129. Epub 2008 Apr 8. Bioinformatics. 2008. PMID: 18397895 - rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure.
Gautier M, Vitalis R. Gautier M, et al. Bioinformatics. 2012 Apr 15;28(8):1176-7. doi: 10.1093/bioinformatics/bts115. Epub 2012 Mar 7. Bioinformatics. 2012. PMID: 22402612 - synbreed: a framework for the analysis of genomic prediction data using R.
Wimmer V, Albrecht T, Auinger HJ, Schön CC. Wimmer V, et al. Bioinformatics. 2012 Aug 1;28(15):2086-7. doi: 10.1093/bioinformatics/bts335. Epub 2012 Jun 10. Bioinformatics. 2012. PMID: 22689388 - Software solutions for the livestock genomics SNP array revolution.
Nicolazzi EL, Biffani S, Biscarini F, Orozco Ter Wengel P, Caprera A, Nazzicari N, Stella A. Nicolazzi EL, et al. Anim Genet. 2015 Aug;46(4):343-53. doi: 10.1111/age.12295. Epub 2015 Apr 23. Anim Genet. 2015. PMID: 25907889 Review. - "Mitochondrial Toolbox" - A Review of Online Resources to Explore Mitochondrial Genomics.
Cappa R, de Campos C, Maxwell AP, McKnight AJ. Cappa R, et al. Front Genet. 2020 May 8;11:439. doi: 10.3389/fgene.2020.00439. eCollection 2020. Front Genet. 2020. PMID: 32457801 Free PMC article. Review.
Cited by
- Adaptive markers distinguish North and South Pacific Albacore amid low population differentiation.
Vaux F, Bohn S, Hyde JR, O'Malley KG. Vaux F, et al. Evol Appl. 2021 Feb 23;14(5):1343-1364. doi: 10.1111/eva.13202. eCollection 2021 May. Evol Appl. 2021. PMID: 34025772 Free PMC article. - Genome-wide SNPs in the spiny lobster Panulirus homarus reveal a hybrid origin for its subspecies.
Farhadi A, Jeffs AG, Lavery SD. Farhadi A, et al. BMC Genomics. 2022 Nov 12;23(1):750. doi: 10.1186/s12864-022-08984-w. BMC Genomics. 2022. PMID: 36368918 Free PMC article. - Low impact of different SNP panels from two building-loci pipelines on RAD-Seq population genomic metrics: case study on five diverse aquatic species.
Casanova A, Maroso F, Blanco A, Hermida M, Ríos N, García G, Manuzzi A, Zane L, Verissimo A, García-Marín JL, Bouza C, Vera M, Martínez P. Casanova A, et al. BMC Genomics. 2021 Mar 2;22(1):150. doi: 10.1186/s12864-021-07465-w. BMC Genomics. 2021. PMID: 33653268 Free PMC article. - Loss of genetic variation and ancestral sex determination system in North American northern pike characterized by whole-genome resequencing.
Johnson HA, Rondeau EB, Sutherland BJG, Minkley DR, Leong JS, Whitehead J, Despins CA, Gowen BE, Collyard BJ, Whipps CM, Farrell JM, Koop BF. Johnson HA, et al. G3 (Bethesda). 2024 Oct 7;14(10):jkae183. doi: 10.1093/g3journal/jkae183. G3 (Bethesda). 2024. PMID: 39115373 Free PMC article. - Ecological characteristics and in situ genetic associations for yield-component traits of wild Miscanthus from eastern Russia.
Clark LV, Dzyubenko E, Dzyubenko N, Bagmet L, Sabitov A, Chebukin P, Johnson DA, Kjeldsen JB, Petersen KK, Jørgensen U, Yoo JH, Heo K, Yu CY, Zhao H, Jin X, Peng J, Yamada T, Sacks EJ. Clark LV, et al. Ann Bot. 2016 Oct 1;118(5):941-955. doi: 10.1093/aob/mcw137. Ann Bot. 2016. PMID: 27451985 Free PMC article.
References
- Aulchenko YS, et al. Genabel: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. - PubMed
- Clayton D, Leung H-T. An R package for analysis of whole-genome association studies. Hum. Hered. 2007;64:45–51. - PubMed
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. - PubMed
- Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources