Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses - PubMed (original) (raw)
Comparative Study
. 2011 Nov;77(21):7663-8.
doi: 10.1128/AEM.00289-11. Epub 2011 Sep 16.
Affiliations
- PMID: 21926223
- PMCID: PMC3209148
- DOI: 10.1128/AEM.00289-11
Comparative Study
Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses
Kyoung-Ho Kim et al. Appl Environ Microbiol. 2011 Nov.
Abstract
Investigation of viruses in the environment often requires the amplification of viral DNA before sequencing of viral metagenomes. In this study, two of the most widely used amplification methods, the linker amplified shotgun library (LASL) and multiple displacement amplification (MDA) methods, were applied to a sample from the seawater surface. Viral DNA was extracted from viruses concentrated by tangential flow filtration and amplified by these two methods. 454 pyrosequencing was used to read the metagenomic sequences from different libraries. The resulting taxonomic classifications of the viruses, their functional assignments, and assembly patterns differed substantially depending on the amplification method. Only double-stranded DNA viruses were retrieved from the LASL, whereas most sequences in the MDA library were from single-stranded DNA viruses, and double-stranded DNA viral sequences were minorities. Thus, the two amplification methods reveal different aspects of viral diversity.
Figures
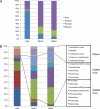
Fig. 1.
Classification of metagenomic sequences from the three libraries based on biological groups (A) and viral genome types and viral families (B) from BLASTX analysis. Among total reads analyzed in the LASL, MDAH, and MDAX libraries, only 14.2%, 6.0%, and 9.3%, respectively, showed hits against the NCBI nonredundant protein database (A) and only 9.8%, 6.2%, and 9.8%, respectively, against the CAMERA viral protein database (B).
Similar articles
- Amplification of uncultured single-stranded DNA viruses from rice paddy soil.
Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y, Jeon CO, Oh HM, Bae JW. Kim KH, et al. Appl Environ Microbiol. 2008 Oct;74(19):5975-85. doi: 10.1128/AEM.01275-08. Epub 2008 Aug 15. Appl Environ Microbiol. 2008. PMID: 18708511 Free PMC article. - Analysis of a viral metagenomic library from 200 m depth in Monterey Bay, California constructed by direct shotgun cloning.
Steward GF, Preston CM. Steward GF, et al. Virol J. 2011 Jun 9;8:287. doi: 10.1186/1743-422X-8-287. Virol J. 2011. PMID: 21651822 Free PMC article. - Comparative metagenomics: natural populations of induced prophages demonstrate highly unique, lower diversity viral sequences.
McDaniel LD, Rosario K, Breitbart M, Paul JH. McDaniel LD, et al. Environ Microbiol. 2014 Feb;16(2):570-85. doi: 10.1111/1462-2920.12184. Epub 2013 Jul 23. Environ Microbiol. 2014. PMID: 23879711 - A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics.
Rosario K, Duffy S, Breitbart M. Rosario K, et al. Arch Virol. 2012 Oct;157(10):1851-71. doi: 10.1007/s00705-012-1391-y. Epub 2012 Jul 4. Arch Virol. 2012. PMID: 22760663 Review. - Barcoding of Plant Viruses with Circular Single-Stranded DNA Based on Rolling Circle Amplification.
Jeske H. Jeske H. Viruses. 2018 Aug 31;10(9):469. doi: 10.3390/v10090469. Viruses. 2018. PMID: 30200312 Free PMC article. Review.
Cited by
- Going viral: next-generation sequencing applied to phage populations in the human gut.
Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Reyes A, et al. Nat Rev Microbiol. 2012 Sep;10(9):607-17. doi: 10.1038/nrmicro2853. Epub 2012 Aug 6. Nat Rev Microbiol. 2012. PMID: 22864264 Free PMC article. Review. - Antibiotics in feed induce prophages in swine fecal microbiomes.
Allen HK, Looft T, Bayles DO, Humphrey S, Levine UY, Alt D, Stanton TB. Allen HK, et al. mBio. 2011 Nov 29;2(6):e00260-11. doi: 10.1128/mBio.00260-11. Print 2011. mBio. 2011. PMID: 22128350 Free PMC article. - Membrane vesicles in sea water: heterogeneous DNA content and implications for viral abundance estimates.
Biller SJ, McDaniel LD, Breitbart M, Rogers E, Paul JH, Chisholm SW. Biller SJ, et al. ISME J. 2017 Feb;11(2):394-404. doi: 10.1038/ismej.2016.134. Epub 2016 Nov 8. ISME J. 2017. PMID: 27824343 Free PMC article. - Quality control implementation for universal characterization of DNA and RNA viruses in clinical respiratory samples using single metagenomic next-generation sequencing workflow.
Bal A, Pichon M, Picard C, Casalegno JS, Valette M, Schuffenecker I, Billard L, Vallet S, Vilchez G, Cheynet V, Oriol G, Trouillet-Assant S, Gillet Y, Lina B, Brengel-Pesce K, Morfin F, Josset L. Bal A, et al. BMC Infect Dis. 2018 Oct 29;18(1):537. doi: 10.1186/s12879-018-3446-5. BMC Infect Dis. 2018. PMID: 30373528 Free PMC article. - Choice of assembly software has a critical impact on virome characterisation.
Sutton TDS, Clooney AG, Ryan FJ, Ross RP, Hill C. Sutton TDS, et al. Microbiome. 2019 Jan 28;7(1):12. doi: 10.1186/s40168-019-0626-5. Microbiome. 2019. PMID: 30691529 Free PMC article.
References
- Breitbart M., et al. 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159:367–373 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources