Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex - PubMed (original) (raw)
Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex
Kimihisa Yoshida et al. Proc Natl Acad Sci U S A. 2011.
Abstract
So far, only a few of the interactions between the ≈ 30 nucleoporins comprising the modular structure of the nuclear pore complex have been defined at atomic resolution. Here we report the crystal structure, at 2.6 Å resolution, of a heterotrimeric complex, composed of fragments of three cytoplasmically oriented nucleoporins of yeast: Nup82, Nup116, and Nup159. Our data show that the Nup82 fragment, representing more than the N-terminal half of the molecule, folds into an extensively decorated, seven-bladed β-propeller that forms the centerpiece of this heterotrimeric complex and anchors both a C-terminal fragment of Nup116 and the C-terminal tail of Nup159. Binding between Nup116 and Nup82 is mutually reinforced via two loops, one emanating from the Nup82 β-propeller and the other one from the β-sandwich fold of Nup116, each contacting binding pockets in their counterparts. The Nup82-Nup159 interaction occurs through an amphipathic α-helix of Nup159, which is cradled in a large hydrophobic groove that is generated from several large surface decorations of the Nup82 β-propeller. Although Nup159 and Nup116 fragments bind to the Nup82 β-propeller in close vicinity, there are no direct contacts between them, consistent with the noncooperative binding that was detected biochemically. Extensive mutagenesis delineated hot-spot residues for these interactions. We also showed that the Nup82 β-propeller binds to other yeast Nup116 family members, Nup145N, Nup100 and to the mammalian homolog, Nup98. Notably, each of the three nucleoporins contains additional nuclear pore complex binding sites, distinct from those that were defined here in the heterotrimeric Nup82•Nup159•Nup116 complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
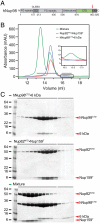
Assembly of a Nup82NTD•Nup159T•Nup116CTD heterotrimer. (A) Domain organization of yeast Nup82, Nup116, and Nup159 (1). Bars denote the fragments that were used for heterotrimer assembly and crystallization: Nup82 N-terminal domain (NTD, blue); Nup116 C-terminal domain (CTD, green); and Nup159 C-terminal tail (T, red). GLEBS, binding site for the RNA export factor Gle2; DID, dynein light chain interacting domain. (B) Analysis by size exclusion chromatography coupled to multiangle light scattering. The differential refractive indices of Nup116CTD (red), Nup82NTD•Nup159T (blue), and Nup82NTD•Nup159T•Nup116CTD (green) are plotted against the elution volumes from a Superdex 200 10/300 GL gel filtration column (GE Healthcare) with dots indicating molar masses.
Fig. 2.
Structural overview of the Nup82NTD•Nup159T•Nup116CTD complex. (A_–_C) Ribbon representation from various angles. Nup82NTD (blue) folds into a β-propeller with various noncanonical insertions highlighted in yellow (3D4A or “FGL” loop), gray (4CD), and orange (6CD); Nup116CTD (green) is a β-sandwich with a β6-αB connector (“K-loop”) indicated in magenta; Nup159T (red) folds into an amphipathic α-helix. Dotted lines represent disordered regions. (D) Schematic representation of the β-propeller of Nup82NTD. Prominent insertions and secondary structure elements are labeled. The asterisk denotes the N-terminal region that fits into the crevice between blades 1 and 7, replacing the canonical Velcro closure observed in most β-propellers.
Fig. 3.
Close-up view of the Nup82-Nup116 and Nup82-Nup159 interfaces. Nup82NTD interacts with Nup116CTD via two adjacent sites. (A) One site is formed by the 3D4A “FGL loop” of Nup82NTD, which binds to a groove of Nup116CTD between helix αB and strand β5. (B) The other site is formed by the “K-loop” of Nup116CTD, K1063 of which binds to a Nup82 pocket with an aspartate (D204) at its bottom and a hydrophobic bracelet at its entry. (C) Nup159T binds to a groove in Nup82NTD that is formed by the 4CD and 6CD insertions and blade 5. Color code as in Fig. 2.
Fig. 4.
Human Nup98CTD binds to yeast Nup82NTD. (A) Domain organization of human Nup98 (1, 18) indicating the C-terminal domain (CTD, green) and the auto-proteolytic 6-kDa protein (gray), as well as the auto-proteolytic cleavage site (red arrow). (B) Size exclusion chromatography analysis of purified yeast Nup82NTD•Nup159T (blue), human Nup98CTD•6-kDa (red) and a mixture (green) in an ≈1∶2 ratio. Note formation of a trimeric Nup82NTD•Nup159T•hNup98CTD complex leads to displacement of the 6-kDa protein (inset). (C) Fractions, indicated by gray bars in (B), of hNup98CTD•6-kDa (top), Nup82NTD•Nup159T (center), and their mixture (bottom) were analyzed by SDS-PAGE; proteins are indicated on the right and molecular weight standards on the left.
Fig. 5.
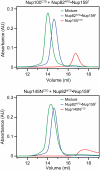
Binding of yeast paralogs of Nup116CTD to Nup82NTD. Interaction analysis between Nup82NTD•Nup159T and the yeast Nup116CTD homologs (A) Nup100CTD and (B) Nup145NCTD. Size exclusion chromatography analysis of Nup82NTD•Nup159T (blue), Nup100CTD (red) and Nup145NCTD (red), and their mixtures (green).
Fig. 6.
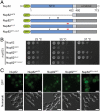
In vivo analysis of Nup82 mutants. (A) Domain organization of the GFP-labeled Nup82 constructs, colored as in Fig. 1_A_. The black and red arrows indicate the positions of the DFY and LILLF mutations, respectively. (B) Yeast growth analysis using a _nup82_Δ strain transformed with the indicated GFP-Nup82 constructs. 10-fold serial dilutions were spotted on SD-Leu plates and grown for 2–3 d at the indicated temperatures. The combination of the DFY and LILLF mutations in one mutant (Nup82DFY–LILLF) has the same effect on growth as the deletion of the entire NTD (Nup82ΔNTD). (C) In vivo localization of the GFP-Nup82 constructs at 37 °C. Both the Nup82DFY–LILLF and the Nup82ΔNTD mutants fail to localize to the nuclear rim. The scale bars represent 5 μm.
Similar articles
- Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex.
Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A. Stuwe T, et al. J Mol Biol. 2012 Jun 22;419(5):330-46. doi: 10.1016/j.jmb.2012.03.024. Epub 2012 Apr 2. J Mol Biol. 2012. PMID: 22480613 Free PMC article. - Structure of a yeast Dyn2-Nup159 complex and molecular basis for dynein light chain-nuclear pore interaction.
Romes EM, Tripathy A, Slep KC. Romes EM, et al. J Biol Chem. 2012 May 4;287(19):15862-73. doi: 10.1074/jbc.M111.336172. Epub 2012 Mar 12. J Biol Chem. 2012. PMID: 22411995 Free PMC article. - Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Sampathkumar P, Kim SJ, Manglicmot D, Bain KT, Gilmore J, Gheyi T, Phillips J, Pieper U, Fernandez-Martinez J, Franke JD, Matsui T, Tsuruta H, Atwell S, Thompson DA, Emtage JS, Wasserman SR, Rout MP, Sali A, Sauder JM, Almo SC, Burley SK. Sampathkumar P, et al. Proteins. 2012 Aug;80(8):2110-6. doi: 10.1002/prot.24102. Epub 2012 Jun 4. Proteins. 2012. PMID: 22544723 Free PMC article. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review.
Cited by
- Evidence for the robustness of protein complexes to inter-species hybridization.
Leducq JB, Charron G, Diss G, Gagnon-Arsenault I, Dubé AK, Landry CR. Leducq JB, et al. PLoS Genet. 2012;8(12):e1003161. doi: 10.1371/journal.pgen.1003161. Epub 2012 Dec 27. PLoS Genet. 2012. PMID: 23300466 Free PMC article. - Crystal structure of the N-terminal domain of Nup358/RanBP2.
Kassube SA, Stuwe T, Lin DH, Antonuk CD, Napetschnig J, Blobel G, Hoelz A. Kassube SA, et al. J Mol Biol. 2012 Nov 9;423(5):752-65. doi: 10.1016/j.jmb.2012.08.026. Epub 2012 Sep 7. J Mol Biol. 2012. PMID: 22959972 Free PMC article. - Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - Evolutionary divergence of the nuclear pore complex from fungi to metazoans.
Chopra K, Bawaria S, Chauhan R. Chopra K, et al. Protein Sci. 2019 Mar;28(3):571-586. doi: 10.1002/pro.3558. Epub 2018 Dec 24. Protein Sci. 2019. PMID: 30488506 Free PMC article. - Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex.
Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K. Onischenko E, et al. Cell. 2017 Nov 2;171(4):904-917.e19. doi: 10.1016/j.cell.2017.09.033. Epub 2017 Oct 12. Cell. 2017. PMID: 29033133 Free PMC article.
References
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. - PubMed
- Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. - PubMed
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–254. - PubMed
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases