Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus - PubMed (original) (raw)
Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus
Katrien Vandersteegen et al. PLoS One. 2011.
Abstract
The increasing antibiotic resistance in bacterial populations requires alternatives for classical treatment of infectious diseases and therefore drives the renewed interest in phage therapy. Methicillin resistant Staphylococcus aureus (MRSA) is a major problem in health care settings and live-stock breeding across the world. This research aims at a thorough microbiological, genomic, and proteomic characterization of S. aureus phage ISP, required for therapeutic applications. Host range screening of a large batch of S. aureus isolates and subsequent fingerprint and DNA microarray analysis of the isolates revealed a substantial activity of ISP against 86% of the isolates, including relevant MRSA strains. From a phage therapy perspective, the infection parameters and the frequency of bacterial mutations conferring ISP resistance were determined. Further, ISP was proven to be stable in relevant in vivo conditions and subcutaneous as well as nasal and oral ISP administration to rabbits appeared to cause no adverse effects. ISP encodes 215 gene products on its 138,339 bp genome, 22 of which were confirmed as structural proteins using tandem electrospray ionization-mass spectrometry (ESI-MS/MS), and shares strong sequence homology with the 'Twort-like viruses'. No toxic or virulence-associated proteins were observed. The microbiological and molecular characterization of ISP supports its application in a phage cocktail for therapeutic purposes.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Dendrogram of the 22 identified S. aureus and S. haemolyticus genotypes.
The dendrogram was generated based on rep-PCR patterns using a 98% similarity level. For each genotype, the representative strain and its fingerprint, the constituting isolates and the ISP sensitivity are indicated.
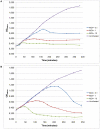
Figure 2. Infection curves of Staphylococcus phage ISP.
Three independent experiments of a non-infected culture and cultures infected with MOI 0.1, 1 and 10 of S. aureus phage propagation strain 47 (A) and S. aureus subsp. aureus Rosenbach ATCC 6538 (B) are shown. Standard deviations are indicated.
Figure 3. Summary of the bio-informatical analysis of the phage ISP genome.
(A) Representation of the genome of phage ISP. Predicted proteins are indicated as bars, putative promoters are depicted as arrows and putative terminators as hair pins and the 4 tRNA's are indicated in light blue. The DNA polymerase and the endolysin, both interrupted by endonucleases, are colored red and orange respectively. Genes which were not annotated in phage G1 are colored purple and genes mistakenly annotated in phage G1 in dark blue, additional genes in phage ISP are framed in red. (B, C and D) Pairwise DNA homology of ISP and G1, ISP and K and ISP and Twort, compared using a sliding window of 50 bp.
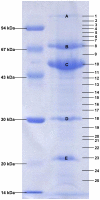
Figure 4. Coomassie G-250 stained SDS-PAGE gel of the structural proteome of phage ISP.
On the left a low molecular weight marker is shown. The numbers on the right correspond to the analyzed gel pieces of which the identified proteins are given in Table 1. Clearly visible protein bands are indicated with capital letters.
Similar articles
- Correlation of Host Range Expansion of Therapeutic Bacteriophage Sb-1 with Allele State at a Hypervariable Repeat Locus.
Sergueev KV, Filippov AA, Farlow J, Su W, Kvachadze L, Balarjishvili N, Kutateladze M, Nikolich MP. Sergueev KV, et al. Appl Environ Microbiol. 2019 Oct 30;85(22):e01209-19. doi: 10.1128/AEM.01209-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31492663 Free PMC article. - Characterization of vB_SauM-fRuSau02, a Twort-Like Bacteriophage Isolated from a Therapeutic Phage Cocktail.
Leskinen K, Tuomala H, Wicklund A, Horsma-Heikkinen J, Kuusela P, Skurnik M, Kiljunen S. Leskinen K, et al. Viruses. 2017 Sep 14;9(9):258. doi: 10.3390/v9090258. Viruses. 2017. PMID: 28906479 Free PMC article. - Identification of gene clusters associated with host adaptation and antibiotic resistance in Chinese Staphylococcus aureus isolates by microarray-based comparative genomics.
Li H, Zhao C, Chen H, Zhang F, He W, Wang X, Wang Q, Yang R, Zhou D, Wang H. Li H, et al. PLoS One. 2013;8(1):e53341. doi: 10.1371/journal.pone.0053341. Epub 2013 Jan 7. PLoS One. 2013. PMID: 23308197 Free PMC article. - Overview of the risks of Staphylococcus aureus infections and their control by bacteriophages and bacteriophage-encoded products.
Rai A, Khairnar K. Rai A, et al. Braz J Microbiol. 2021 Dec;52(4):2031-2042. doi: 10.1007/s42770-021-00566-4. Epub 2021 Jul 12. Braz J Microbiol. 2021. PMID: 34251609 Free PMC article. Review. - Bacteriophage Therapy for Staphylococcus Aureus Infections: A Review of Animal Models, Treatments, and Clinical Trials.
Plumet L, Ahmad-Mansour N, Dunyach-Remy C, Kissa K, Sotto A, Lavigne JP, Costechareyre D, Molle V. Plumet L, et al. Front Cell Infect Microbiol. 2022 Jun 17;12:907314. doi: 10.3389/fcimb.2022.907314. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35782148 Free PMC article. Review.
Cited by
- Stability of bacteriophages in burn wound care products.
Merabishvili M, Monserez R, van Belleghem J, Rose T, Jennes S, De Vos D, Verbeken G, Vaneechoutte M, Pirnay JP. Merabishvili M, et al. PLoS One. 2017 Jul 27;12(7):e0182121. doi: 10.1371/journal.pone.0182121. eCollection 2017. PLoS One. 2017. PMID: 28750102 Free PMC article. - ε2-Phages Are Naturally Bred and Have a Vastly Improved Host Range in Staphylococcus aureus over Wild Type Phages.
Sáez Moreno D, Visram Z, Mutti M, Restrepo-Córdoba M, Hartmann S, Kremers AI, Tišáková L, Schertler S, Wittmann J, Kalali B, Monecke S, Ehricht R, Resch G, Corsini L. Sáez Moreno D, et al. Pharmaceuticals (Basel). 2021 Apr 2;14(4):325. doi: 10.3390/ph14040325. Pharmaceuticals (Basel). 2021. PMID: 33918287 Free PMC article. - Structure and transformation of bacteriophage A511 baseplate and tail upon infection of Listeria cells.
Guerrero-Ferreira RC, Hupfeld M, Nazarov S, Taylor NM, Shneider MM, Obbineni JM, Loessner MJ, Ishikawa T, Klumpp J, Leiman PG. Guerrero-Ferreira RC, et al. EMBO J. 2019 Feb 1;38(3):e99455. doi: 10.15252/embj.201899455. Epub 2019 Jan 2. EMBO J. 2019. PMID: 30606715 Free PMC article. - Selection and characterization of a candidate therapeutic bacteriophage that lyses the Escherichia coli O104:H4 strain from the 2011 outbreak in Germany.
Merabishvili M, De Vos D, Verbeken G, Kropinski AM, Vandenheuvel D, Lavigne R, Wattiau P, Mast J, Ragimbeau C, Mossong J, Scheres J, Chanishvili N, Vaneechoutte M, Pirnay JP. Merabishvili M, et al. PLoS One. 2012;7(12):e52709. doi: 10.1371/journal.pone.0052709. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23285164 Free PMC article. - Insights into Gene Transcriptional Regulation of Kayvirus Bacteriophages Obtained from Therapeutic Mixtures.
Arroyo-Moreno S, Buttimer C, Bottacini F, Chanishvili N, Ross P, Hill C, Coffey A. Arroyo-Moreno S, et al. Viruses. 2022 Mar 17;14(3):626. doi: 10.3390/v14030626. Viruses. 2022. PMID: 35337034 Free PMC article.
References
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. 2344
- Monecke S, Jatzwauk L, Weber S, Slickers P, Ehricht R. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin Microbiol Infect. 2008;14:534–545. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical