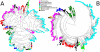Toward an efficient method of identifying core genes for evolutionary and functional microbial phylogenies - PubMed (original) (raw)
Toward an efficient method of identifying core genes for evolutionary and functional microbial phylogenies
Nicola Segata et al. PLoS One. 2011.
Abstract
Microbial community metagenomes and individual microbial genomes are becoming increasingly accessible by means of high-throughput sequencing. Assessing organismal membership within a community is typically performed using one or a few taxonomic marker genes such as the 16S rDNA, and these same genes are also employed to reconstruct molecular phylogenies. There is thus a growing need to bioinformatically catalog strongly conserved core genes that can serve as effective taxonomic markers, to assess the agreement among phylogenies generated from different core gene, and to characterize the biological functions enriched within core genes and thus conserved throughout large microbial clades. We present a method to recursively identify core genes (i.e. genes ubiquitous within a microbial clade) in high-throughput from a large number of complete input genomes. We analyzed over 1,100 genomes to produce core gene sets spanning 2,861 bacterial and archaeal clades, ranging in size from one to >2,000 genes in inverse correlation with the α-diversity (total phylogenetic branch length) spanned by each clade. These cores are enriched as expected for housekeeping functions including translation, transcription, and replication, in addition to significant representations of regulatory, chaperone, and conserved uncharacterized proteins. In agreement with previous manually curated core gene sets, phylogenies constructed from one or more of these core genes agree with those built using 16S rDNA sequence similarity, suggesting that systematic core gene selection can be used to optimize both comparative genomics and determination of microbial community structure. Finally, we examine functional phylogenies constructed by clustering genomes by the presence or absence of orthologous gene families and show that they provide an informative complement to standard sequence-based molecular phylogenies.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Evolutionarily conserved core gene sets calculated using the NCBI Taxonomy as a guide tree.
In the circular cladograms each node represents a taxon, ranging from phyla (internal) to genera, species, and in some cases strains and sub-strains (external) with yellow leaf nodes representing taxa for which a complete genome is available. Core genes for higher-level clades are indicated by color (from black to red) proportional to the logarithm of the number of core genes, and white circles represent clades without cores. A) The full tree of core gene sets, representing 2,861 clades and 1,107 sequenced organisms. This tree includes the Bacteria and Archaea and results in cores that are functionally enriched for housekeeping genes including basic DNA and RNA operations. B) The core gene tree limited for visual clarity to the family. Note that our core gene discovery algorithm reflects for differences in phylogenetic depth and includes fewer core genes for broader clades spanning greater diversity.
Figure 2. Hierarchical clustering of organisms based on the COG gene families present in their genomes.
Heat maps represent individual COG orthologous gene family abundances (columns) for each taxon (rows); absent gene families are white, single copy families are black, and multicopy families red. The resulting functional phylogeny represents a type of phylogenetic profiling and clearly highlights functional characteristics specific to groups of organisms independent of their evolutionary relatedness. In many cases, evolutionary and functional similarities are highly correlated; three representative clusters are enlarged in green boxes and the three most abundant COGs for each cluster are reported. Note that the absence of the bacterial ribosome and DNA maintenance machinery from the Archaea is readily apparent from such data, and that the Enterobacteriaceae and Lactobacillales both include striking large clusters of strongly conserved uncharacterized genes.
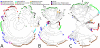
Figure 3. Contrasting a functional phylogeny built using shared gene families with a 16S gene sequence phylogeny.
1,107 sequenced microbes clustered using (A) the cooccurence of COG orthologous gene families (see Figure 2) or (B) 16S gene sequence similarity (using Muscle and FastTree [35]). Phyla from the NCBI Taxonomy are indicated by color. While overall organismal similarities are maintained at both the functional and sequence levels, the two phylogenies provide distinct perspectives on organismal relatedness. Some clades like the Archaea and Firmicutes form very distinct sub-trees, whereas others like Bacteroidetes/Chlorobi, Actinobacteria and Cyanobacteria show high 16S similarity relative to their functional similarity.
Figure 4. Firmicutes phylogenies obtained using functional clustering, 16S rRNA gene sequence, or core gene sequence.
Colored leaves represent taxonomic orders. Trees generated using (A) functional similarity of COGs as detailed in Figure 2, (B) 16S rRNA gene similarity, and (C) core gene sequence similarity (for excinuclease ABC subunit A, the only core gene found for Firmicutes). Note that while overall tree structure is comparable for the three methods, functional phylogeny and the excinuclease ABC subunit A core gene sequence correctly assign organize the Bacillus cereus group with other Bacillus and Geobacillus genera within the Bacillaceae.
References
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, et al. Toward automatic reconstruction of a highly resolved tree of life. Science (New York, NY) 2006;311:1283–1287. - PubMed
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources