A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae) - PubMed (original) (raw)
A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae)
Lenka Caisová et al. BMC Evol Biol. 2011.
Abstract
Background: The second Internal Transcriber Spacer (ITS2) is a fast evolving part of the nuclear-encoded rRNA operon located between the 5.8S and 28S rRNA genes. Based on crossing experiments it has been proposed that even a single Compensatory Base Change (CBC) in helices 2 and 3 of the ITS2 indicates sexual incompatibility and thus separates biological species. Taxa without any CBC in these ITS2 regions were designated as a 'CBC clade'. However, in depth comparative analyses of ITS2 secondary structures, ITS2 phylogeny, the origin of CBCs, and their relationship to biological species have rarely been performed. To gain 'close-up' insights into ITS2 evolution, (1) 86 sequences of ITS2 including secondary structures have been investigated in the green algal order Ulvales (Chlorophyta, Viridiplantae), (2) after recording all existing substitutions, CBCs and hemi-CBCs (hCBCs) were mapped upon the ITS2 phylogeny, rather than merely comparing ITS2 characters among pairs of taxa, and (3) the relation between CBCs, hCBCs, CBC clades, and the taxonomic level of organisms was investigated in detail.
Results: High sequence and length conservation allowed the generation of an ITS2 consensus secondary structure, and introduction of a novel numbering system of ITS2 nucleotides and base pairs. Alignments and analyses were based on this structural information, leading to the following results: (1) in the Ulvales, the presence of a CBC is not linked to any particular taxonomic level, (2) most CBC 'clades' sensu Coleman are paraphyletic, and should rather be termed CBC grades. (3) the phenetic approach of pairwise comparison of sequences can be misleading, and thus, CBCs/hCBCs must be investigated in their evolutionary context, including homoplasy events (4) CBCs and hCBCs in ITS2 helices evolved independently, and we found no evidence for a CBC that originated via a two-fold hCBC substitution.
Conclusions: Our case study revealed several discrepancies between ITS2 evolution in the Ulvales and generally accepted assumptions underlying ITS2 evolution as e.g. the CBC clade concept. Therefore, we developed a suite of methods providing a critical 'close-up' view into ITS2 evolution by directly tracing the evolutionary history of individual positions, and we caution against a non-critical use of the ITS2 CBC clade concept for species delimitation.
Figures
Figure 1
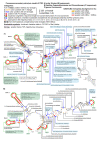
Consensus secondary structure models of ITS2 in the Ulvales. A) Consensus ITS2 diagram based on 86 sequences covering five families (Kornmanniaceae, Bolbocoleonaceae, Ulvaceae, Capsosiphonaceae and Gomontiaceae). B) ITS2 consensus of the Capsosiphonaceae and Gomontiaceae (41 sequences analyzed), showing extremely high conservation. Nucleotide letters shown in both ITS2 diagrams (A, B) refer to the most frequently occurring character states among the analyzed taxa, obtained via 70% majority rule consensus sequences. Positions without 'dominant' character state among the investigated Ulvales were integrated as circles, or flagged as expansion segments. Invariable positions were drawn in black, whereas for variable positions, the conservation/variability level was quantified by the number of evolutionary changes during the diversification of the Ulvales, and indicated by various colors (nucleotides and/or circles in ochre, blue, or red). 129 positions were present in all studied Ulvales, and were used for a 'universal' numbering system of ITS2 positions. The 'non-universal' positions were labeled with subscript numbers, combined with the previous 'universal' position number. Gray shades indicate the conserved parts of helices 2 and 3 [6]. Several comments in the Figure refer to non-homoplasious CBCs (black frames), inserted positions characteristic for selected taxa, and the length variation of expansion segments. The CBC/hCBC homoplasy background in the ITS2 diagram (A) is indicated by ochre/blue/pale pink shades with the restriction to positions with CBCs/hCBCs/CBCs+hCBCs respectively. C) Simplified ITS2 diagram of Kornmannia displaying a unique, additional helix between helices 3 and 4.
Figure 2
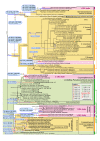
Evolution of CBCs in paired ITS2 nucleotides mapped upon the ITS2 phylogeny of the Ulvales. All compensatory base changes (CBCs) accompanied by appropriate Helix (H) and numbers of positions (by specific nucleotide numbers) were linked to the nodes/branches where they evolved. CBCs that occurred in the conserved regions of helices 2 and 3 (H2+3_CBCs) were shown in bold and in larger font size and their corresponding branches were depicted in bold as well. Branches in blue are characterized by CBCs and hCBCs, whereas branches in red color have CBC support exclusively. Only those taxa, which formed a terminal, monophyletic clade and were not differentiated by any CBC in the conserved parts of ITS2 helices (H2+3_CBCs), were here designated as a CBC clade, and were highlighted in pink background color. In contrast, taxa lacking distinguishing H2+3_CBCs, which formed non-monophyletic assemblies in the phylogenetic tree, were designated as 'CBC grades', and shaded in orange color. Note that all CBC grades contained nested CBC clades. Typically, a CBC clade/grade can be traced back to a common ancestor (basal branch) characterized by synapomorphic H2+3_CBCs, except for one 'plesiomorphic CBC grade' (green color) characterized merely by shared plesiomorphies in helices 2 and 3 of ITS2. CBCs either evolved as unique (non-homoplasious; NHS) or homoplasious synapomorphies (HS). CBCs were homoplasious due to parallelisms (PAR 1-7), convergences (CONV 1-3) and/or reversals (REV 1-2), and all these changes were mapped upon the tree (encircled numbers). The tree topology was based on 152 aligned ITS2 characters from 86 taxa analyzed by maximum likelihood (ML). The branch separating the Capsosiphonaceae/Gomontiaceae from the remaining Ulvales was used for rooting the tree. Four interrupted branches have been graphically reduced to 50% or 75% of the original length. Significances at branches from left to right are bootstrap percentages (ML, NJ, and MP) and Bayesian posterior probabilities. Newly determined sequences (12) are in bold (for accession numbers see Additional file 7). Taxa/strains with hash mark (#) were also analyzed in the 18S rRNA phylogeny (Additional file 2).
Figure 3
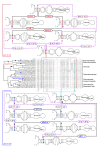
Evolution of hCBCs in ITS2 base pairs in the Ulvales. Hemi-compensatory base changes (hCBCs) referring to conserved parts of helices 2 and 3 were shown in bold and in larger font size and their corresponding branches were illustrated in bold. Branches in red are characterized by hCBCs and CBCs, whereas branches in green color have hCBC support exclusively. Encircled numbers were used to indicate all hCBC-type parallelisms (hPAR 1-14) and reversals (hREV 1-6); hCBC-type convergences were not found in the ITS2 of the Ulvales.
Figure 4
Phenetic versus phylogenetic approach of species delimitation of two taxa of Acrochaete. A) Phenetic approach, i.e. pair wise comparison without consideration of the plesiomorphic status of the base pair 21/40 in the conserved region of Helix2 (H2) revealed that A. viridis and A. heteroclada differ by only one hCBC (A-U vs. G-U, respectively). In contrast, B) a phylogenetic approach taking the ancestral status (G-C) of the respective base pair into consideration resulted in the difference of one CBC (G-C → A-U) + one hCBC (G-C → G-U) between these two taxa. Whereas the phenetic approach would,- according to the CBC clade concept, regard A. viridis and A. heteroclada to belong to a single CBC clade (and potentially the same species), the phylogenetic approach showed A. viridis and A. heteroclada as two separate species. Base pair 21/40 as well as its plesiomorphic status (both in gray boxes) are mapped on the branches; and the evolving CBC/hCBC are indicated by blue dashed arrows.
Figure 5
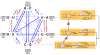
Parallelisms, convergences, and reversals in ITS2 base pairs, as shown by selected examples. All three cases of homoplasious changes occurred in ITS2 pairs 54/121 and 55/120 in the basal part of Helix 3 in the Ulvales. For all taxa in the simplified phylogram (derived from Figure 2), sequences of the basal part of Helix 3 are presented as an alignment, and linked to secondary structure diagrams highlighting the homoplasious character evolution of base pair 54/121 (in red color, above alignment) and pair 55/120 (in blue color, below alignment). Dashed arrows in violet were used to indicate the evolutionary direction of homoplasious changes. In the phylogram, branches/clades in which homoplasious substitutions occurred were marked by red/blue circles. Homoplasious changes were abbreviated as before (Figures 2, 3).
Figure 6
Occurrence/frequency of substitutions of ITS2 base pairs, and homoplasious changes, mapped upon ITS2 helices. Occurrence and frequency of all compensatory (i.e. CBCs, hCBCs) and non-compensatory substitutions of ITS2 base pairs, and homoplasious changes (parallelisms, convergences, reversals), mapped upon ITS2 helices. Eight invariant base pairs are in bold; the conserved parts of helices 2 and 3 are in gray. Base pairs displaying CBCs + hCBCs are indicated in pink, pairs evolved exclusively via CBCs are in blue, and pairs developed solely via hCBCs are drawn in red. Non-compensating substitutions (N-N ⇔ N×N) were especially frequent in homoplasious positions of Helix 1 and in two pairs of Helix 3 (55/120, 62/114). For Helix 1 and 3 base pairs, the total number of non-compensatory changes cannot be estimated precisely due to their high substitution frequency.
Figure 7
Diagram showing evolutionary changes of base pairs, and their frequency/occurrence in ITS2 of the Ulvales. All possible evolutionary changes between canonical (C-G, A-U) and 'wobble' (G-U) base pairs in RNA molecules, and their frequency of occurrence in the entire ITS2 are illustrated. Base pairs are given in 5'-3' orientation, referring to their placement in a helix. Arrows indicate the evolutionary direction of substitutions. ITS2 changes, which were found in the Ulvales are indicated by bold arrows, accompanied by frequencies (encircled numbers), whereas changes that were not existent in the analyzed taxa (frequency 0) are shown as thin arrows. CBCs are shown in blue, hCBCs are in red. Obviously, hCBCs (especially the G-C ⇔ G-U type) occurred more frequently than CBCs. From the frequency of base pairs (number in black boxes) and its percentage (number below black boxes) it is evident that there was a strong selection towards the GC/CG category. Note that the given frequencies of base pairs are confined to extant taxa. The illustration of Helix 1 with pair 8/11 highlighted refers to the hypothesis that a CBC of the U-A ⇔ C-G type may have evolved via two consecutive hCBC steps (see Results). Note, however, that the taxa shown are unrelated, indicated by the simplified trees, which provide no support for this hypothesis.
Similar articles
- A consensus secondary structure of ITS2 in the chlorophyta identified by phylogenetic reconstruction.
Caisová L, Marin B, Melkonian M. Caisová L, et al. Protist. 2013 Jul;164(4):482-96. doi: 10.1016/j.protis.2013.04.005. Epub 2013 Jun 13. Protist. 2013. PMID: 23770573 - Evolutionary diversification indicated by compensatory base changes in ITS2 secondary structures in a complex fungal species, Rhizoctonia solani.
Ahvenniemi P, Wolf M, Lehtonen MJ, Wilson P, German-Kinnari M, Valkonen JP. Ahvenniemi P, et al. J Mol Evol. 2009 Aug;69(2):150-63. doi: 10.1007/s00239-009-9260-3. Epub 2009 Jul 16. J Mol Evol. 2009. PMID: 19609478 - A consensus secondary structure of ITS2 for the diatom Order Cymatosirales (Mediophyceae, Bacillariophyta) and reappraisal of the order based on DNA, morphology, and reproduction.
Samanta B, Ehrman JM, Kaczmarska I. Samanta B, et al. Mol Phylogenet Evol. 2018 Dec;129:117-129. doi: 10.1016/j.ympev.2018.08.014. Epub 2018 Aug 25. Mol Phylogenet Evol. 2018. PMID: 30153502 - ITS2 sequence-structure analysis in phylogenetics: a how-to manual for molecular systematics.
Schultz J, Wolf M. Schultz J, et al. Mol Phylogenet Evol. 2009 Aug;52(2):520-3. doi: 10.1016/j.ympev.2009.01.008. Mol Phylogenet Evol. 2009. PMID: 19489124 Review. - Phylogenetic Utility of rRNA ITS2 Sequence-Structure under Functional Constraint.
Zhang W, Tian W, Gao Z, Wang G, Zhao H. Zhang W, et al. Int J Mol Sci. 2020 Sep 3;21(17):6395. doi: 10.3390/ijms21176395. Int J Mol Sci. 2020. PMID: 32899108 Free PMC article. Review.
Cited by
- New deep-sea species of Aborjinia (Nematoda, Leptosomatidae) from the North-Western Pacific: an integrative taxonomy and phylogeny.
Zograf JK, Semenchenko AA, Mordukhovich VV. Zograf JK, et al. Zookeys. 2024 Jan 17;1189:231-256. doi: 10.3897/zookeys.1189.111825. eCollection 2024. Zookeys. 2024. PMID: 38282715 Free PMC article. - Applying an internal transcribed spacer as a single molecular marker to differentiate between Tetraselmis and Chlorella species.
Fathy WA, Techen N, Elsayed KNM, Essawy EA, Tawfik E, Alwutayd KM, Abdelhameed MS, Hammouda O, Ross SA. Fathy WA, et al. Front Microbiol. 2023 Aug 23;14:1228869. doi: 10.3389/fmicb.2023.1228869. eCollection 2023. Front Microbiol. 2023. PMID: 37680531 Free PMC article. - Characterizing nrDNA ITS1, 5.8S and ITS2 secondary structures and their phylogenetic utility in the legume tribe Hedysareae with special reference to Hedysarum.
Nafisi H, Kaveh A, Kazempour-Osaloo S. Nafisi H, et al. PLoS One. 2023 Apr 12;18(4):e0283847. doi: 10.1371/journal.pone.0283847. eCollection 2023. PLoS One. 2023. PMID: 37043495 Free PMC article. - Hyaloscypha gabretae and Hyaloscypha gryndleri spp. nov. (Hyaloscyphaceae, Helotiales), two new mycobionts colonizing conifer, ericaceous and orchid roots.
Vohník M, Figura T, Réblová M. Vohník M, et al. Mycorrhiza. 2022 Jan;32(1):105-122. doi: 10.1007/s00572-021-01064-z. Epub 2022 Jan 14. Mycorrhiza. 2022. PMID: 35028741 - Phylogenetic Reassessment, Taxonomy, and Biogeography of Codinaea and Similar Fungi.
Réblová M, Kolařík M, Nekvindová J, Réblová K, Sklenář F, Miller AN, Hernández-Restrepo M. Réblová M, et al. J Fungi (Basel). 2021 Dec 20;7(12):1097. doi: 10.3390/jof7121097. J Fungi (Basel). 2021. PMID: 34947079 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources