Isolated NIBPL missense mutations that cause Cornelia de Lange syndrome alter MAU2 interaction - PubMed (original) (raw)
doi: 10.1038/ejhg.2011.175. Epub 2011 Sep 21.
Melanie Hullings, María Concepcion Gil-Rodríguez, Christopher T Fincher, Mark B Mallozzi, Elizabeth Loy, Melanie Albrecht, Maninder Kaur, Janusz Limon, Abhinav Rampuria, Dinah Clark, Antonie Kline, Andreas Dalski, Juliane Eckhold, Andreas Tzschach, Raoul Hennekam, Gabriele Gillessen-Kaesbach, Jolanta Wierzba, Ian D Krantz, Matthew A Deardorff, Frank J Kaiser
Affiliations
- PMID: 21934712
- PMCID: PMC3283175
- DOI: 10.1038/ejhg.2011.175
Isolated NIBPL missense mutations that cause Cornelia de Lange syndrome alter MAU2 interaction
Diana Braunholz et al. Eur J Hum Genet. 2012 Mar.
Erratum in
- Eur J Hum Genet. 2012 Mar;20(3):366
Abstract
Cornelia de Lange syndrome (CdLS; or Brachmann-de Lange syndrome) is a dominantly inherited congenital malformation disorder with features that include characteristic facies, cognitive delays, growth retardation and limb anomalies. Mutations in nearly 60% of CdLS patients have been identified in NIPBL, which encodes a regulator of the sister chromatid cohesion complex. NIPBL, also known as delangin, is a homolog of yeast and amphibian Scc2 and C. elegans PQN-85. Although the exact mechanism of NIPBL function in sister chromatid cohesion is unclear, in vivo yeast and C. elegans experiments and in vitro vertebrate cell experiments have demonstrated that NIPBL/Scc2 functionally interacts with the MAU2/Scc4 protein to initiate loading of cohesin onto chromatin. To test the significance of this model in the clinical setting of CdLS, we fine-mapped the NIBPL-MAU2 interaction domain and tested the functional significance of missense mutations and variants in NIPBL and MAU2 identified in these minimal domains in a cohort of patients with CdLS. We demonstrate that specific novel mutations at the N-terminus of the MAU2-interacting domain of NIBPL result in markedly reduced MAU2 binding, although we appreciate no consistent clinical difference in the small group of patients with these mutations. These data suggest that factors in addition to MAU2 are essential in determining the clinical features and severity of CdLS.
Figures
Figure 1
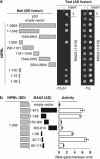
Localization of NIPBL–MAU2 interaction. (a) NIPBL fragments used in confirmation and localization of NIPBL binding to MAU2 are demonstrated in gray to the left. Amino-acid residues included are indicated. The right panel demonstrates yeast two-hybrid colony assays, indicating interaction-dependent growth on tryptophan-, leucine- and histidine-deficient media (−T/L/H) of the positive control p53 with SV40TAg (T) in the uppermost row. Interaction-independent growth on tryptophan- and leucine-deficient plates is indicated to the right. NIBPL clones were tested with empty (-) and MAU2 containing AD fusion vectors. (b) The left panel depicts the NIPBL (aa 1–300) and MAU2 deletion constructs used for liquid _β_-galactosidase assay. The right panel indicates the interaction of the NIPBL/delangin fragment (1–300) and the different MAU2 protein fragments by liquid _β_-galactosidase assay.
Figure 2
The effect of NIPBL mutations on NIPBL/delangin–MAU2 interaction. (a) NIPBL/delangin motifs include a glutamine-rich domain (GLN), nuclear localization signal (NLS) and HEAT-repeat motifs. The minimal MAU-2 interaction fragment used (aa 1–38) and relative positions of mutations tested are demonstrated. (b) Constructs tested are indicated in the right panel and interaction data from mammalian two-hybrid assays are indicated in relative luciferase units for each CdLS mutation tested at the left. Significance of difference in activity of the G15R and P29Q versus wild-type (WT) are indicated. (c) Western blotting of NIPBL-BD 1–300 fragments with missense mutations from two-hybrid assays, using anti-GAL4 (DBD) antibody (Santa Cruz). All proteins are expressed at similar levels.
Figure 3
Clinical features of patients. Frontal and profile face photos and photos of hands are shown for the patients described. Images for an individual patient are outlined by a black box, with the patient's mutation indicated.
Similar articles
- MAU2 and NIPBL Variants Impair the Heterodimerization of the Cohesin Loader Subunits and Cause Cornelia de Lange Syndrome.
Parenti I, Diab F, Gil SR, Mulugeta E, Casa V, Berutti R, Brouwer RWW, Dupé V, Eckhold J, Graf E, Puisac B, Ramos F, Schwarzmayr T, Gines MM, van Staveren T, van IJcken WFJ, Strom TM, Pié J, Watrin E, Kaiser FJ, Wendt KS. Parenti I, et al. Cell Rep. 2020 May 19;31(7):107647. doi: 10.1016/j.celrep.2020.107647. Cell Rep. 2020. PMID: 32433956 - Clinical study and genetic analysis of Cornelia de Lange syndrome caused by a novel MAU2 gene variant in a Chinese boy.
Peng Y, Zhu Y, Wu L, Deng F. Peng Y, et al. Mol Genet Genomic Med. 2024 Jan;12(1):e2318. doi: 10.1002/mgg3.2318. Epub 2023 Nov 14. Mol Genet Genomic Med. 2024. PMID: 37962004 Free PMC article. - The effect of Nipped-B-like (Nipbl) haploinsufficiency on genome-wide cohesin binding and target gene expression: modeling Cornelia de Lange syndrome.
Newkirk DA, Chen YY, Chien R, Zeng W, Biesinger J, Flowers E, Kawauchi S, Santos R, Calof AL, Lander AD, Xie X, Yokomori K. Newkirk DA, et al. Clin Epigenetics. 2017 Aug 25;9:89. doi: 10.1186/s13148-017-0391-x. eCollection 2017. Clin Epigenetics. 2017. PMID: 28855971 Free PMC article. - Cornelia de Lange Syndrome and the link between chromosomal function, DNA repair and developmental gene regulation.
Strachan T. Strachan T. Curr Opin Genet Dev. 2005 Jun;15(3):258-64. doi: 10.1016/j.gde.2005.04.005. Curr Opin Genet Dev. 2005. PMID: 15917200 Review. - Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome.
Mannini L, Cucco F, Quarantotti V, Krantz ID, Musio A. Mannini L, et al. Hum Mutat. 2013 Dec;34(12):1589-96. doi: 10.1002/humu.22430. Epub 2013 Sep 16. Hum Mutat. 2013. PMID: 24038889 Free PMC article. Review.
Cited by
- Mutant cohesin affects RNA polymerase II regulation in Cornelia de Lange syndrome.
Mannini L, C Lamaze F, Cucco F, Amato C, Quarantotti V, Rizzo IM, Krantz ID, Bilodeau S, Musio A. Mannini L, et al. Sci Rep. 2015 Nov 19;5:16803. doi: 10.1038/srep16803. Sci Rep. 2015. PMID: 26581180 Free PMC article. - Roberts syndrome: A deficit in acetylated cohesin leads to nucleolar dysfunction.
Xu B, Lu S, Gerton JL. Xu B, et al. Rare Dis. 2014 Jan 21;2:e27743. doi: 10.4161/rdis.27743. eCollection 2014. Rare Dis. 2014. PMID: 25054091 Free PMC article. - The Emerging Role of Cohesin in the DNA Damage Response.
Litwin I, Pilarczyk E, Wysocki R. Litwin I, et al. Genes (Basel). 2018 Nov 28;9(12):581. doi: 10.3390/genes9120581. Genes (Basel). 2018. PMID: 30487431 Free PMC article. Review. - Independent mechanisms recruit the cohesin loader protein NIPBL to sites of DNA damage.
Bot C, Pfeiffer A, Giordano F, Manjeera DE, Dantuma NP, Ström L. Bot C, et al. J Cell Sci. 2017 Mar 15;130(6):1134-1146. doi: 10.1242/jcs.197236. Epub 2017 Feb 6. J Cell Sci. 2017. PMID: 28167679 Free PMC article. - Cornelia de Lange syndrome with NIPBL mutation and mosaic Turner syndrome in the same individual.
Wierzba J, Gil-Rodríguez MC, Polucha A, Puisac B, Arnedo M, Teresa-Rodrigo ME, Winnicka D, Hegardt FG, Ramos FJ, Limon J, Pié J. Wierzba J, et al. BMC Med Genet. 2012 Jun 7;13:43. doi: 10.1186/1471-2350-13-43. BMC Med Genet. 2012. PMID: 22676896 Free PMC article.
References
- Kline AD, Krantz ID, Sommer A, et al. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A. 2007;143:1287–1296. - PubMed
- Ireland M. Cornelia de Lange syndrome: clinical features, common complications and long-term prognosis. Curr Paediatr. 1996;6:69–73.
- Kline AD, Stanley C, Belevich J, Brodsky K, Barr M, Jackson LG. Developmental data on individuals with the Brachmann-de Lange syndrome. Am J Med Genet. 1993;47:1053–1058. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials