Molecular organization of vomeronasal chemoreception - PubMed (original) (raw)
Molecular organization of vomeronasal chemoreception
Yoh Isogai et al. Nature. 2011.
Abstract
The vomeronasal organ (VNO) has a key role in mediating the social and defensive responses of many terrestrial vertebrates to species- and sex-specific chemosignals. More than 250 putative pheromone receptors have been identified in the mouse VNO, but the nature of the signals detected by individual VNO receptors has not yet been elucidated. To gain insight into the molecular logic of VNO detection leading to mating, aggression or defensive responses, we sought to uncover the response profiles of individual vomeronasal receptors to a wide range of animal cues. Here we describe the repertoire of behaviourally and physiologically relevant stimuli detected by a large number of individual vomeronasal receptors in mice, and define a global map of vomeronasal signal detection. We demonstrate that the two classes (V1R and V2R) of vomeronasal receptors use fundamentally different strategies to encode chemosensory information, and that distinct receptor subfamilies have evolved towards the specific recognition of certain animal groups or chemical structures. The association of large subsets of vomeronasal receptors with cognate, ethologically and physiologically relevant stimuli establishes the molecular foundation of vomeronasal information coding, and opens new avenues for further investigating the neural mechanisms underlying behaviour specificity.
Figures
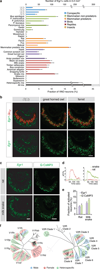
Figure 1. Egr1 expression is robustly induced by pheromone-evoked VNO neuronal activation
Female CD-1 mice were exposed to clean or male mouse bedding and their VNOs analyzed for expression of various immediate early genes (IEGs). a, In situ hybridization with RNA probes to Arc, c-Fos, c-Jun, Egr1, FosB, and Nr4a1 b, Numbers of IEG positive cells after bedding exposure (10 sections per VNO, n=3 animals). Error bars show s.e.m. c, TrpC2, a cation channel involved in VNO signal transduction is required for Egr1 induction. Female TrpC2+/− or TrpC2−/− mice were exposed to male conspecific bedding and Egr1 expression was visualized in the VNO. Bar, 100 µm.
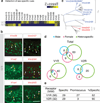
Figure 2. Widespread activation of VNO receptors by conspecific and heterospecific cues
a, Survey of ethologically relevant vomeronasal stimuli. Vomeronasal neural activation upon exposure to conspecific and heterospecific cues was visualized by Egr1 induction and quantified. Detection of female cues by males is designated as ♀(♂). Unless specified, female mice were used. Mixed heterospecific cues activated Egr1 in significantly more cells than individual stimuli (p<0.01, two-tailed t-test). Co-exposure to heterospecific and conspecific stimuli (all mix, n=6) resulted in significantly more _Egr1_ positive cells (p<0.05, two-tailed t-test). **b**, Widespread activation of VNO neurons by conspecific and heterospecific cues. Shown are in situ hybridization with probes against _Gαi2_ (red) and _Egr1_ (green). **c**, Comparison between _Egr1_ and G-CaMP3-evoked signal in response to rat or milk snake chemosignals. G-CaMP3 images are 10 sec averages of ΔF frames within stimulus period. **d**, Differential response profiles of neurons to rat or snake signals. Stimuli were perfused from 20 sec to 60 sec. **e**, Quantitative comparison between _Egr1_ and G-CaMP3 evoked-signals. The percentage of activated cells identified by G-CaMP3 (n = 356 cells for rat stimuli, n = 566 cells for snake stimuli, 9 VNO slices from 3 animals) among those responsive to 40 mM KCl was plotted in the graph. Data for _Egr1_ was taken from the Fig. 2a. The difference between _Egr1_ and G-CaMP3 was not statistically significant (p>0.1, two tailed t-test). f, Clade-level maps of V1R (left) and V2R (right) activation show distinct clade specificity for male, female or heterospecific cues. Hatched patterns indicate response to multiple types of cues. Error bars are in s.e.m. Scale bars show 100 µm.
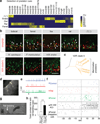
Figure 3. Receptor repertoires to sex-specific cues
a, b, Male and female mouse cues are each detected by a specific subset of V1Rs and V2Rs. a, Heat maps representing the co-localization between Egr1 and representative vomeronasal receptor genes (yellow 100%, blue 0% overlap). b, In situ hybridization of Egr1 (green) and individual receptors (red), with arrows marking co-localization of Egr1 and receptor signals. The scale bars represent 100 µm. c, Clade organization of V2Rs detecting male or female cues. d, Receptors detecting male, female, and heterospecific cues are largely distinct. e, V1Rs and V2Rs display distinct specificity. The table shows the number of receptors that detect unique types of cues (specific) versus multiple types (promiscuous) among the following categories: male, female, mammalian non-predator, mammalian predator, reptile, and avian predator.
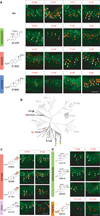
Figure 4. Receptor repertoires to heterospecific cues
a, b, Predator cues are detected by a specific subset of V1Rs and V2Rs. a, Heat map representing the co-localization between Egr1 and representative vomeronasal receptor genes (yellow 100%, blue 0% overlap). b, In situ hybridization of Egr1 (green) and vomeronasal receptors (red), with arrows marking co-localization of Egr1 and receptor signals. Bar, 100 µm. b,c, Mammalian predator cues commonly activate V2R clade 5 receptors. Due to high homology among V2R clade 5 genes, the Vmn2r30, 33, 34, 39 probes detect multiple receptors. d, Fluorescence image showing a patched V1rh7-YFP neuron. e, Loose-patch recordings of a V1rh7-YFP neuron. The arrow indicates perfusion start. f, Spike raster for three different VNO neurons showing responses of a V1rh7-YFP neuron and a V1Rh7-YFP negative neuron. The stimulus perfusion started at −30 sec and lasted 20 seconds. g, h, Rat bedding (arrow) elicits robust avoidance behaviors in control TrpC2+/− mice, but significantly less in TrpC2−/− mice lacking VNO activity. *** indicates p<0.0001 (two tailed Student’s t-Test). Error bars show s.e.m. (TrpC2+/−, n=13, TrpC2−/−, n=17).
Figure 5. Sulfated steroids detection by V1Rs
a, V1Ref, and V1Rjk clade specific probes (red) co-localize with Egr1 (green) after VNO stimulation by a mix of steroids containing the glucocorticoid Q1570, the estrogen E1050, and the androgen A7864. Each of these compounds elicits activity in distinct populations of vomeronasal neurons (V1re2, V1re6, V1rf3, and V1rj2), also represented in the molecular tree of V1R receptors (b). c, The three distinct estradiols activate both V1rf3 and V1rj2 while the estriol only activates V1rf3. d, The sulfate group position in pregnenes determine the specificity of ligand detection by V1re2 and V1re6. Bar, 100 µm.
Comment in
- Sensory systems: Charting vomeronasal receptor function.
Whalley K. Whalley K. Nat Rev Neurosci. 2011 Oct 5;12(11):618. doi: 10.1038/nrn3123. Nat Rev Neurosci. 2011. PMID: 21971068 No abstract available.
Similar articles
- Chemoreception: identifying friends and foes.
Koh TW, Carlson JR. Koh TW, et al. Curr Biol. 2011 Dec 20;21(24):R998-9. doi: 10.1016/j.cub.2011.10.038. Curr Biol. 2011. PMID: 22192835 - Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes.
Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, Ogawa S, Zufall F, Mombaerts P. Del Punta K, et al. Nature. 2002 Sep 5;419(6902):70-4. doi: 10.1038/nature00955. Nature. 2002. PMID: 12214233 - Vomeronasal Receptors in Vertebrates and the Evolution of Pheromone Detection.
Silva L, Antunes A. Silva L, et al. Annu Rev Anim Biosci. 2017 Feb 8;5:353-370. doi: 10.1146/annurev-animal-022516-022801. Epub 2016 Nov 28. Annu Rev Anim Biosci. 2017. PMID: 27912243 Review. - Pheromone detection by mammalian vomeronasal neurons.
Zufall F, Kelliher KR, Leinders-Zufall T. Zufall F, et al. Microsc Res Tech. 2002 Aug 1;58(3):251-60. doi: 10.1002/jemt.10152. Microsc Res Tech. 2002. PMID: 12203702 Review. - Functional Overexpression of Vomeronasal Receptors Using a Herpes Simplex Virus Type 1 (HSV-1)-Derived Amplicon.
Stein B, Alonso MT, Zufall F, Leinders-Zufall T, Chamero P. Stein B, et al. PLoS One. 2016 May 19;11(5):e0156092. doi: 10.1371/journal.pone.0156092. eCollection 2016. PLoS One. 2016. PMID: 27195771 Free PMC article.
Cited by
- Interspecies sex and taste.
Koh TW, Carlson JR. Koh TW, et al. Cell. 2013 Jul 3;154(1):20-1. doi: 10.1016/j.cell.2013.06.015. Cell. 2013. PMID: 23827670 Free PMC article. - Cyclic Regulation of Sensory Perception by a Female Hormone Alters Behavior.
Dey S, Chamero P, Pru JK, Chien MS, Ibarra-Soria X, Spencer KR, Logan DW, Matsunami H, Peluso JJ, Stowers L. Dey S, et al. Cell. 2015 Jun 4;161(6):1334-44. doi: 10.1016/j.cell.2015.04.052. Cell. 2015. PMID: 26046438 Free PMC article. - Tuning properties and dynamic range of type 1 vomeronasal receptors.
Haga-Yamanaka S, Ma L, Yu CR. Haga-Yamanaka S, et al. Front Neurosci. 2015 Jul 14;9:244. doi: 10.3389/fnins.2015.00244. eCollection 2015. Front Neurosci. 2015. PMID: 26236183 Free PMC article. - Mouse alarm pheromone shares structural similarity with predator scents.
Brechbühl J, Moine F, Klaey M, Nenniger-Tosato M, Hurni N, Sporkert F, Giroud C, Broillet MC. Brechbühl J, et al. Proc Natl Acad Sci U S A. 2013 Mar 19;110(12):4762-7. doi: 10.1073/pnas.1214249110. Epub 2013 Mar 4. Proc Natl Acad Sci U S A. 2013. PMID: 23487748 Free PMC article. - Bacterial MgrB peptide activates chemoreceptor Fpr3 in mouse accessory olfactory system and drives avoidance behaviour.
Bufe B, Teuchert Y, Schmid A, Pyrski M, Pérez-Gómez A, Eisenbeis J, Timm T, Ishii T, Lochnit G, Bischoff M, Mombaerts P, Leinders-Zufall T, Zufall F. Bufe B, et al. Nat Commun. 2019 Oct 25;10(1):4889. doi: 10.1038/s41467-019-12842-x. Nat Commun. 2019. PMID: 31653840 Free PMC article.
References
- Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nature Reviews Neuroscience. 2003;4:551–562. - PubMed
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. - PubMed
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials