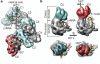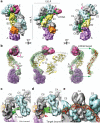Structures of the RNA-guided surveillance complex from a bacterial immune system - PubMed (original) (raw)
Structures of the RNA-guided surveillance complex from a bacterial immune system
Blake Wiedenheft et al. Nature. 2011.
Abstract
Bacteria and archaea acquire resistance to viruses and plasmids by integrating short fragments of foreign DNA into clustered regularly interspaced short palindromic repeats (CRISPRs). These repetitive loci maintain a genetic record of all prior encounters with foreign transgressors. CRISPRs are transcribed and the long primary transcript is processed into a library of short CRISPR-derived RNAs (crRNAs) that contain a unique sequence complementary to a foreign nucleic-acid challenger. In Escherichia coli, crRNAs are incorporated into a multisubunit surveillance complex called Cascade (CRISPR-associated complex for antiviral defence), which is required for protection against bacteriophages. Here we use cryo-electron microscopy to determine the subnanometre structures of Cascade before and after binding to a target sequence. These structures reveal a sea-horse-shaped architecture in which the crRNA is displayed along a helical arrangement of protein subunits that protect the crRNA from degradation while maintaining its availability for base pairing. Cascade engages invading nucleic acids through high-affinity base-pairing interactions near the 5' end of the crRNA. Base pairing extends along the crRNA, resulting in a series of short helical segments that trigger a concerted conformational change. This conformational rearrangement may serve as a signal that recruits a trans-acting nuclease (Cas3) for destruction of invading nucleic-acid sequences.
© 2011 Macmillan Publishers Limited. All rights reserved
Figures
Figure 1. Structure of the Cascade complex from E. coli
a The CRISPR system in E. coli K12 (Cse-type) consists of eight cas genes and a downstream CRISPR locus. casA to casE are members of large gene families, referred to as cse1, cse2, cse4, cas5e and cse3, respectively,. The CRISPR consists of a series of 29-nucleotide repeats (black diamonds) separated by 32-nucleotide spacer sequences (green cylinders). CasE (magenta) is an endoribonuclease that specifically binds to a stable stem–loop in the CRISPR RNA repeat and cleaves 8 nucleotides away from the spacer sequence in the 5′ direction,,. b, Cascade assembles into a sea-horse-shaped architecture where the crRNA (green) is positioned along a helical arrangement of six CasC subunits (C1–6). The helical spine is capped at its ends by two prominent features that represent the head (E, CasE) and tail (A, CasA) of the sea-horse anatomy. D, CasD. c, Cascade consists of unequal numbers of Cas proteins and a crRNA (CasA1B2C6D1E1crRNA1). The first five CasC subunits (C1–5) are structurally similar, whereas CasC6 is distinct. B1, CasB1; B2, CasB2.
Figure 2. Programmed capping of the CasC helix
a, C1–5 form a right-handed helix with a pitch of 135Å. The two domains of each CasC subunit are referred to as proximal and distal, relative to the helical axis of the CasC subunits. The different conformation of C6 (red) relative to the other CasC subunits interrupts the helical symmetry (black arrow). b, The crRNA is positioned along a contiguous groove on the concave surface of the C1–5 helix. The 5′ end of the crRNA forms a hook-like structure that interacts with C6. This interaction correlates with the distinct conformation of C6 and the termination of the helix. Although the proximal domains of C5 and C6 have the same orientation, the distal domain of C6 is rotated by ~160° relative to the other CasC subunits (black arrow). The centre of rotation is indicated by a black dot.
Figure 3. Target binding triggers a concerted conformational change
a, Structure of Cascade bound to a 32-nucleotide target RNA complementary to the crRNA spacer sequence, at a resolution of ~9 Å. b, Removing the CasC subunits reveals significant structural differences between the unbound and target-bound structures. CasE, CasA and the crRNA from the unbound structure are shown as grey volumes. The same subunits from the target bound complex are shown in magenta, green mesh and purple mesh, respectively. Models of the CasB crystal structure are shown docked into the cryo-EM map before (grey) and after (yellow) target binding. Four or five base pairs of double-stranded RNA (red) fit with high fidelity into the crRNA density. On target binding, CasE rotates by ~15° (magenta arrow), both CasB subunits move ~17Å along the concave surface of the CasC backbone (yellow arrow) and CasA rotates by ~30° (purple arrow). The purple asterisks indicate the positions of the four-helix bundle on CasA before and after target binding. c, In the unbound structure, CasC6 is rotated out of the CasC helix, exposing the 5′ region of the crRNA (double-headed arrow indicates density for the single-stranded crRNA). d, Additional density corresponding to the target nucleic acid is clearly visible in the target-bound complex (double-headed arrows). Base pairing in the crRNA spacer disrupts the hook-like structure at the 5′ end of the crRNA, and a difference map reveals a significant loss of resolvable density in the distal domain of C6 following target binding (red mesh and arrow). e, The short segments of double-stranded helices are connected by short, non-helical regions located at pinch points of the CasC subunits (white arrows).
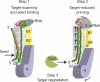
Figure 4. A model for pathogen surveillance and signalling by Cascade
Efficient surveillance and detection of invading nucleic acids is mediated by base pairing in the seed sequence (nucleotides 1–5, 7–8) of the crRNA. Duplex formation may proceed in the 3′ direction (four curved black arrows), resulting in a series of short helical duplexes that shorten the crRNA; this in turn causes a concerted conformational change in CasA, CasB and CasE (coloured arrows) that coincides with a disruption of the 5′ hook and results in a decrease in resolvable density for the distal domain of C6. Cascade binds single-stranded and double-stranded substrates. Here we depict the target as a single strand for simplicity.
Similar articles
- CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference.
Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. Hochstrasser ML, et al. Proc Natl Acad Sci U S A. 2014 May 6;111(18):6618-23. doi: 10.1073/pnas.1405079111. Epub 2014 Apr 18. Proc Natl Acad Sci U S A. 2014. PMID: 24748111 Free PMC article. - The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers.
Luo ML, Jackson RN, Denny SR, Tokmina-Lukaszewska M, Maksimchuk KR, Lin W, Bothner B, Wiedenheft B, Beisel CL. Luo ML, et al. Nucleic Acids Res. 2016 Sep 6;44(15):7385-94. doi: 10.1093/nar/gkw421. Epub 2016 May 12. Nucleic Acids Res. 2016. PMID: 27174938 Free PMC article. - Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli.
Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Jackson RN, et al. Science. 2014 Sep 19;345(6203):1473-9. doi: 10.1126/science.1256328. Epub 2014 Aug 7. Science. 2014. PMID: 25103409 Free PMC article. - Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems.
Richter C, Chang JT, Fineran PC. Richter C, et al. Viruses. 2012 Oct 19;4(10):2291-311. doi: 10.3390/v4102291. Viruses. 2012. PMID: 23202464 Free PMC article. Review. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review.
Cited by
- Crystal structure of the largest subunit of a bacterial RNA-guided immune complex and its role in DNA target binding.
Mulepati S, Orr A, Bailey S. Mulepati S, et al. J Biol Chem. 2012 Jun 29;287(27):22445-9. doi: 10.1074/jbc.C112.379503. Epub 2012 May 23. J Biol Chem. 2012. PMID: 22621933 Free PMC article. - CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3.
Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJ, van der Oost J. Westra ER, et al. Mol Cell. 2012 Jun 8;46(5):595-605. doi: 10.1016/j.molcel.2012.03.018. Epub 2012 Apr 19. Mol Cell. 2012. PMID: 22521689 Free PMC article. - Mechanism of foreign DNA selection in a bacterial adaptive immune system.
Sashital DG, Wiedenheft B, Doudna JA. Sashital DG, et al. Mol Cell. 2012 Jun 8;46(5):606-15. doi: 10.1016/j.molcel.2012.03.020. Epub 2012 Apr 19. Mol Cell. 2012. PMID: 22521690 Free PMC article. - Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli.
van Erp PB, Jackson RN, Carter J, Golden SM, Bailey S, Wiedenheft B. van Erp PB, et al. Nucleic Acids Res. 2015 Sep 30;43(17):8381-91. doi: 10.1093/nar/gkv793. Epub 2015 Aug 3. Nucleic Acids Res. 2015. PMID: 26243775 Free PMC article. - Targeted Transcriptional Repression in Bacteria Using CRISPR Interference (CRISPRi).
Hawkins JS, Wong S, Peters JM, Almeida R, Qi LS. Hawkins JS, et al. Methods Mol Biol. 2015;1311:349-62. doi: 10.1007/978-1-4939-2687-9_23. Methods Mol Biol. 2015. PMID: 25981485 Free PMC article.
References
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. - PubMed
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
- Bolotin A, Ouinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. - PubMed
- Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases