Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts - PubMed (original) (raw)
Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts
Nicolas Cerveau et al. Genome Biol Evol. 2011.
Abstract
Transposable elements (TE) are one of the major driving forces of genome evolution, raising the question of the long-term dynamics underlying their evolutionary success. Long-term TE evolution can readily be reconstructed in eukaryotes, thanks to many degraded copies constituting genomic fossil records of past TE proliferations. By contrast, bacterial genomes usually experience high sequence turnover and short TE retention times, thereby obscuring ancient TE evolutionary patterns. We found that Wolbachia bacterial genomes contain 52-171 insertion sequence (IS) TEs. IS account for 11% of Wolbachia wRi, which is one of the highest IS genomic coverage reported in prokaryotes to date. We show that many IS groups are currently expanding in various Wolbachia genomes and that IS horizontal transfers are frequent among strains, which can explain the apparent synchronicity of these IS proliferations. Remarkably, >70% of Wolbachia IS are nonfunctional. They constitute an unusual bacterial IS genomic fossil record providing direct empirical evidence for a long-term IS evolutionary dynamics following successive periods of intense transpositional activity. Our results show that comprehensive IS annotations have the potential to provide new insights into prokaryote TE evolution and, more generally, prokaryote genome evolution. Indeed, the identification of an important IS genomic fossil record in Wolbachia demonstrates that IS elements are not always of recent origin, contrary to the conventional view of TE evolution in prokaryote genomes. Our results also raise the question whether the abundance of IS fossils is specific to Wolbachia or it may be a general, albeit overlooked, feature of prokaryote genomes.
Figures
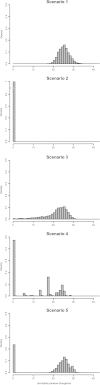
FIG. 1.—
Frequency distribution of simulated pairwise IS divergence in a haploid genome under five models of IS dynamics. Scenario 1: single ancient burst; scenario 2: single recent burst; scenario 3: slow expansion; scenario 4: two independent recent bursts; and scenario 5: ancient and recent bursts. Each distribution represents the pooled pairwise divergence distribution from 23 simulations, each of which has an expected final number of elements equal to the size of 1 of the 23 Wolbachia IS groups comprising at least two alignable IS copies.
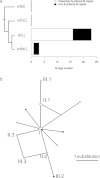
FIG. 2.—
Expansion of the ISWen2 group in the _w_Ri genome. (a) Copy number of the ISWen2 group in four completely sequenced Wolbachia genomes. Branch lengths of the phylogenetic tree are arbitrary. (b) Median-joining network of the 20 full-length ISWen2 copies from the _w_Ri genome. Circles denote IS sequence types (nodes). Nodes discussed in the main text were labeled I, II.1–3, and III.1–3. Node size is proportional to IS copy number: n = 1 for all nodes except nodes I (n = 7) and II.1 (n = 3). Lines denote substitution steps, with a one-step distance being indicated in the lower right corner. Potentially functional and nonfunctional copies are shown in white and black, respectively.
FIG. 3.—
Frequency distribution of pairwise IS nucleotide divergence for four Wolbachia genomes. IS copies from 23 IS groups comprising at least two alignable IS copies are considered (n = 454). The distribution is based on a total of 4,312 pairwise comparisons (169 for _w_Bm, 379 for _w_Mel, 2152 for _w_Pel, and 1612 for _w_Ri).
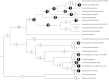
FIG. 4.—
History of IS group acquisitions and losses in 22 Wolbachia strains. The most parsimonious distribution of acquisitions (white circles) and losses (black circles) of 17 IS groups according to the phylogenetic relationships of 22 Wolbachia strains is shown. Numbers of acquisitions and losses are indicated in the circles. Phylogenetic relationships between Wolbachia strains are adapted from Cordaux et al. (2001, and Lo et al. (2007); branch lengths of the phylogenetic tree are arbitrary. Wolbachia strains are named after the host species from which they were isolated.
References
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. - PubMed
- Bichsel M, Barbour AD, Wagner A. The early phase of a bacterial insertion sequence infection. Theor Popul Biol. 2010;78(4):278–288. - PubMed
- Bordenstein SR, Reznikoff WS. Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol. 2005;3(9):688–699. - PubMed
- Bordenstein SR, Wernegreen JJ. Bacteriophage flux in end (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21(10):1981–1991. - PubMed
- Bouchon D, Cordaux R, Grève P. Feminizing Wolbachia and the evolution of sex determination in isopods. In: Bourtzis K, Miller T, editors. Insect Symbiosis. 2008. Insect Symbiosis, Vol. 3. Boca Raton (FL): Taylor and Francis Group LLC. p. 273–294.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous