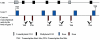DNA methylation: superior or subordinate in the epigenetic hierarchy? - PubMed (original) (raw)
DNA methylation: superior or subordinate in the epigenetic hierarchy?
Bilian Jin et al. Genes Cancer. 2011 Jun.
Abstract
Epigenetic modifications are heritable changes in gene expression not encoded by the DNA sequence. In the past decade, great strides have been made in characterizing epigenetic changes during normal development and in disease states like cancer. However, the epigenetic landscape has grown increasingly complicated, encompassing DNA methylation, the histone code, noncoding RNA, and nucleosome positioning, along with DNA sequence. As a stable repressive mark, DNA methylation, catalyzed by the DNA methyltransferases (DNMTs), is regarded as a key player in epigenetic silencing of transcription. DNA methylation may coordinately regulate the chromatin status via the interaction of DNMTs with other modifications and with components of the machinery mediating those marks. In this review, we will comprehensively examine the current understanding of the connections between DNA methylation and other epigenetic marks and discuss molecular mechanisms of transcriptional repression in development and in carcinogenesis.
Keywords: DNA methylation; DNA methyltransferase; chromatin; epigenetics; histone code.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures
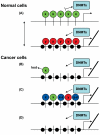
Figure 1.
How the histone code may direct DNA methylation during development and carcinogenesis. (A) During normal development, the transcriptionally activating mark H3K4 me3 (x) blocks or repels DNMTs, whereas the repressive marks H3K9me or H3K27me (y) permit or recruit DNMTs, possibly by direct protein-protein interactions (e.g., EZH2-DNMTs). During carcinogenesis, disruption of the histone code in the form of (B) loss of H3K4me3 (x), (C) substitution of H3K4me3 with H3K9me or H3K27me (y), randomization of marks, aberrant acquisition of a new mark (z), or (D) loss of all histone marks permits or actively induces DNMT recruitment. x = H3K4me3; y = H3K9me or H3K27me; z = other histone mark; bent arrow = transcription start site; lollipops = histone marks; black circles = DNA methylation.
Figure 2.
How the histone code may rely on the DNA methylation machinery for direction. Upon binding to CpG-rich regions, DNMTs may directly recruit HMTs to these domains. During DNA replication, UHRF1 preferentially binds hemimethylated DNA and interacts with/recruits DNMT1 and G9A. PCNA may also have a role in the recruitment process. Methyl-CpG–binding proteins (MBDs) specifically interact with methylated DNA and may form complexes with HMTs such as SETDB1 to direct histone methylation to regions of DNA methylation.
Figure 3.
Intragenic CpG islands (CGIs) function as alternative promoters. CGIs embedded in the body of gene T may function as alternative promoters, whose methylation inversely correlates with the transcriptional levels of their corresponding genes. However, the total average level of intragenic DNA methylation appears to display a positive correlation with the transcription of gene T.
Similar articles
- Epigenetic response to environmental stress: Assembly of BRG1-G9a/GLP-DNMT3 repressive chromatin complex on Myh6 promoter in pathologically stressed hearts.
Han P, Li W, Yang J, Shang C, Lin CH, Cheng W, Hang CT, Cheng HL, Chen CH, Wong J, Xiong Y, Zhao M, Drakos SG, Ghetti A, Li DY, Bernstein D, Chen HS, Quertermous T, Chang CP. Han P, et al. Biochim Biophys Acta. 2016 Jul;1863(7 Pt B):1772-81. doi: 10.1016/j.bbamcr.2016.03.002. Epub 2016 Mar 4. Biochim Biophys Acta. 2016. PMID: 26952936 Free PMC article. - DNMTs and Impact of CpG Content, Transcription Factors, Consensus Motifs, lncRNAs, and Histone Marks on DNA Methylation.
Loaeza-Loaeza J, Beltran AS, Hernández-Sotelo D. Loaeza-Loaeza J, et al. Genes (Basel). 2020 Nov 12;11(11):1336. doi: 10.3390/genes11111336. Genes (Basel). 2020. PMID: 33198240 Free PMC article. Review. - The "Epigenetic Code Replication Machinery", ECREM: a promising drugable target of the epigenetic cell memory.
Bronner C, Chataigneau T, Schini-Kerth VB, Landry Y. Bronner C, et al. Curr Med Chem. 2007;14(25):2629-41. doi: 10.2174/092986707782023244. Curr Med Chem. 2007. PMID: 17979715 Review. - Epigenetic regulation of DNA methyltransferases: DNMT1 and DNMT3B in gliomas.
Rajendran G, Shanmuganandam K, Bendre A, Muzumdar D, Goel A, Shiras A. Rajendran G, et al. J Neurooncol. 2011 Sep;104(2):483-94. doi: 10.1007/s11060-010-0520-2. Epub 2011 Jan 13. J Neurooncol. 2011. PMID: 21229291 - Epigenetic interplay between histone modifications and DNA methylation in gene silencing.
Vaissière T, Sawan C, Herceg Z. Vaissière T, et al. Mutat Res. 2008 Jul-Aug;659(1-2):40-8. doi: 10.1016/j.mrrev.2008.02.004. Epub 2008 Feb 29. Mutat Res. 2008. PMID: 18407786 Review.
Cited by
- 4mCPred-MTL: Accurate Identification of DNA 4mC Sites in Multiple Species Using Multi-Task Deep Learning Based on Multi-Head Attention Mechanism.
Zeng R, Cheng S, Liao M. Zeng R, et al. Front Cell Dev Biol. 2021 May 10;9:664669. doi: 10.3389/fcell.2021.664669. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34041243 Free PMC article. - Epigenetic Therapies in Triple-Negative Breast Cancer: Concepts, Visions, and Challenges.
Lehmann U. Lehmann U. Cancers (Basel). 2024 Jun 7;16(12):2164. doi: 10.3390/cancers16122164. Cancers (Basel). 2024. PMID: 38927870 Free PMC article. Review. - TNFα Induces DNA and Histone Hypomethylation and Pulmonary Artery Smooth Muscle Cell Proliferation Partly via Excessive Superoxide Formation.
Crosswhite P, Sun Z. Crosswhite P, et al. Antioxidants (Basel). 2024 May 31;13(6):677. doi: 10.3390/antiox13060677. Antioxidants (Basel). 2024. PMID: 38929115 Free PMC article. - H3K9 Trimethylation Silences Fas Expression To Confer Colon Carcinoma Immune Escape and 5-Fluorouracil Chemoresistance.
Paschall AV, Yang D, Lu C, Choi JH, Li X, Liu F, Figueroa M, Oberlies NH, Pearce C, Bollag WB, Nayak-Kapoor A, Liu K. Paschall AV, et al. J Immunol. 2015 Aug 15;195(4):1868-82. doi: 10.4049/jimmunol.1402243. Epub 2015 Jul 1. J Immunol. 2015. PMID: 26136424 Free PMC article. - mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis.
Villa E, Sahu U, O'Hara BP, Ali ES, Helmin KA, Asara JM, Gao P, Singer BD, Ben-Sahra I. Villa E, et al. Mol Cell. 2021 May 20;81(10):2076-2093.e9. doi: 10.1016/j.molcel.2021.03.009. Epub 2021 Mar 22. Mol Cell. 2021. PMID: 33756106 Free PMC article.
References
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597-610 - PubMed
Grants and funding
- R01 AA019976/AA/NIAAA NIH HHS/United States
- R01 CA114229/CA/NCI NIH HHS/United States
- R01 CA116028/CA/NCI NIH HHS/United States
- R01 CA116028-06/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous