Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells - PubMed (original) (raw)
. 2011 Sep 23;35(3):400-12.
doi: 10.1016/j.immuni.2011.06.015.
Kenneth J Oestreich, Sang-Jun Ha, Jaikumar Duraiswamy, Rama S Akondy, Erin E West, Zhengyu Wei, Peiyuan Lu, James W Austin, James L Riley, Jeremy M Boss, Rafi Ahmed
Affiliations
- PMID: 21943489
- PMCID: PMC3183460
- DOI: 10.1016/j.immuni.2011.06.015
Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells
Ben Youngblood et al. Immunity. 2011.
Abstract
Functionally exhausted T cells have high expression of the PD-1 inhibitory receptor, and therapies that block PD-1 signaling show promise for resolving chronic viral infections and cancer. By using human and murine systems of acute and chronic viral infections, we analyzed epigenetic regulation of PD-1 expression during CD8(+) T cell differentiation. During acute infection, naive to effector CD8(+) T cell differentiation was accompanied by a transient loss of DNA methylation of the Pdcd1 locus that was directly coupled to the duration and strength of T cell receptor signaling. Further differentiation into functional memory cells coincided with Pdcd1 remethylation, providing an adapted program for regulation of PD-1 expression. In contrast, the Pdcd1 regulatory region was completely demethylated in exhausted CD8(+) T cells and remained unmethylated even when virus titers decreased. This lack of DNA remethylation leaves the Pdcd1 locus poised for rapid expression, potentially providing a signal for premature termination of antiviral functions.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Figure 1
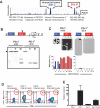
Conserved regions of the PDCD1 promoter contain differentially methylated sites that correlate with PD-1 expression. A) The PDCD1 gene contains several highly conserved regions as determined by the UCSC Genome Browser
(Karolchik et al., 2003). This genomic region contains a CpG island in the human genome as defined by CpG Plot from the European Bioinformatics Institute. The location of the CpG island overlaps with a DNase I hypersensitive region identified by Oestreich et al.(Oestreich et al., 2008). B) Genomic DNA from EL4 (PD-1hi) and A20 (PD-1lo) was digested with either _Hpa_II (H, Methylation sensitive) or _Msp_I (M, non-sensitive) along with _Hind_III (HIII, non-sensitive). A southern blot of the methylation sensitive restriction endonuclease challenge reveals that A20 cells have a methylated promoter. The undigested Hind III fragment is indicated by the arrowhead. The cartoon of the promoter region displays the approximate location of the restriction cut sites. C) Bisulfite sequencing of Conserved Region C (CR-C) and CR-B was performed on genomic DNA from A20 and EL4 cells. Each line represents an individual clone picked for sequencing. Filled circles = methylated cytosine. Open circles = non-methylated cytosine. D) Peripheral blood CD8+ T cells were column purified to >95% purity and stimulated with anti-CD3/CD28 beads for 3 days. Stimulated cells were cultured for an additional 5 days with and without 5-aza-2′-deoxycytidine. Cells were harvested and analyzed by FACS for PD-1 and CD8 expression. E) PD-1 mRNA from the indicated CD8 T cell population were analyzed using qRT-PCR . The values obtained were normalized to 18s rRNA and expressed as fold over the amount of transcript from Day 0 cells.
Figure 2
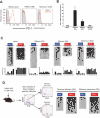
DNA methylation of Pdcd1 CR-C and CR-B is inversely correlated with PD-1 expression in antigen-specific cells from mice acutely infected with LCMV. A) Histogram analysis of PD-1 protein expression on antigen-specific CD8 T cells at 4, 8, and >30 dpi during acute infection with 2*105 pfu of LCMV Armstrong. The red line = LCMV-specific CD8 T cells. The filled gray histogram = naïve CD8 T cells. B) Real-time PCR analysis of PD-1 transcript from purified naïve, day 4 effector, day 8 effector, and memory antigen-specific CD8 T cells. C) Bisulfite sequencing analysis and graphical summary of Pdcd1 CR-C and CR-B from FACS purified naïve, day 4 effector, day 8 effector, and memory cells. D) Bisulfite sequencing of Pdcd1 CR-C and CR-B was performed on genomic DNA from terminal effector and memory precursor CD8 T cell subsets purified from mice at 8 days post infection with 2*105 LCMV Armstrong. Filled circles = methylated cytosine. Each line represents a sequenced clone. Open circles = nonmethylated cytosine. CpG sites 1 – 14 correspond to sites -1280, -1158, -1099, -1069, -986, -778, -672, -667, -636, -612, -535, -496, -491, and -465.
Figure 3
Chronic antigen exposure results in prolonged demethylation of Pdcd1 CR-C and CR-B. A) Histogram analysis of PD-1 protein expression on antigen-specific CD8 T cells during acute infection at 4, 8, and 60 dpi with 2*106 pfu of LCMV clone 13. The red line = LCMV-specific CD8 T cells. The filled gray histogram = naïve CD8 T cells. B) Real-time PCR analysis of PD-1 transcript from purified naïve and exhausted antigen-specific CD8 T cells. C) Bisulfite sequencing analysis and graphical summary of Pdcd1 CR-C and CR-B from FACS purified antigen-specific cells at 8 and >30 dpi from mice infected with LCMV clone 13. D) P14 chimeric mice were infected with 2*105 pfu of LCMV Armstrong, 2*106 LCMV Armstrong, and 2*106 LCMV clone-13. PD-1 expression was measured on the LCMV-specific CD8 T cells harvested at day 8 post infection from the acutely and chronically infected mice. Adoptively transferred LCMV-specific CD8 T cells were purified at 8 days post infection. E) Bisulfite sequencing and collective summary of CpG methylation at all sites in Pdcd1 CR-B was performed on purified Day 8-effector cells. F) Day 40 LCMV-specific cells isolated from LCMV clone 13 infected mice that were treated with GK1.5 to deplete CD4 T cells to achieve full exhaustion. Antigen-specific cells were harvested from the spleen and purified by FACS using. Bisulfite sequencing analysis and graphical summary of CR-C and CR-B from the fully exhausted antigen-specific cells.
Figure 4
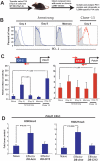
CR-C chromatin is hypersensitive to digestion by DNase I in PD-1hi virus-specific CD8 T cells. A) Chimeric mice were generated by adoptive transfer of P14 transgenic cells. Antigen-specific transgenic cells were harvested from chimeric mice following at 4, 9, and >40 dpi with LCMV clone-13 or Armstrong. Cells were then column purified using a biotinylated antibody to the congenic marker Thy1.1 on the P14 cells and assayed for DNase I hypersensitive regions in the Pdcd1 promoter. Viremia is detectable only in mice infected with clone 13 at 9 dpi vs. mice infected with Armstrong at 9 dpi (Data not shown). B) Histogram panels show PD-1 expression on naïve (shaded gray line), LCMV Armstrong differentiated (open blue line), and LCMV clone-13 differentiated (open red line) P14 LCMV-specific CD8 T cells. C) Relative DNase I hypersensitivity for CR-C and CR-B in the Pdcd1 regulatory region of naïve, day 4-effector, day 9-effector and memory CD8 T cells is plotted. P values were determined using the student T test. Not shown, the P value for the difference in DNase I hypersensitivity of CR-C between naïve and Day 4 effector cells is <0.005. D) Chromatin immunoprecipitation analysis for histone 3 lysine 9 tri-methylation (H3K9me3) and histone 3 lysine 27 tri-methylation (H3K27me3) at Pdcd1 CR-C was performed on chromatin extracted from the purified naïve, D8 effector-armstrong, and D8 effector-clone13 CD8 T cells. The bar graphs were generated from an average of three independent experiments.
Figure 5
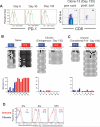
Prolonged antigen exposure results in an inability for antigen-specific CD8 T cells to reinstate the epigenetic program. A) PD-1 expression at day 8, 30 and 135 post LCMV clone-13 infection reveals that PD-1 expression decreases following the clearance of virus. Day 135 pi antigen-specific cells were sorted for methylation analysis. Serum virus titers (PFU/ml) for days 8, 30, and 135 post-infection were 6*104, 6*103, and undetectable respectively. B) Methylation analysis was performed on DNA from antigen-specific cells from naïve and virus-specific cells isolated from clone-13 infected mice at >135 dpi. Bisulfite sequencing analysis and graphical summary of CR-C & B reveals that the promoter is not remethylated in endogenous LCMV-specific CD8 T cells C) Endogenous levels (~200) of transgenic P14 CD8 T cells specific for the LCMV antigen gp33-41 were adoptively transferred into WT B6 mice. Chimeric mice were infected the next day with 2*106 pfu of LCMV clone 13. Bisulfite sequencing was performed on purified adoptively transferred LCMV-specific CD8 T cells 180 dpi. D) P14 Chimeric mice were infected with either LCMV Armstrong or LCMV clone-13. Splenocytes were isolated from infected mice at 6 months pi when PD-1 expression was significantly reduced on antigen-specific cells from clone-13 infected mice. Antigen-specific cells from either immune mice or chronically infected mice were cultured for 0, 6, and 12 hours in media containing the gp33 peptide. The amount of PD-1 expression on the virus-specific CD8 T cells (gated on Thy1.1+ cells) was determined by flow cytometric analysis. LCMV antigen-specific cells from the long-term chronic environment (blue line) produce more PD-1 relative to antigen-specific cells from the acute environment (red line) upon restimulation.
Figure 6
Persistent TCR stimulation results in downregulation of the stem cell isoform of the de novo methyltransferase Dnmt3a. The relative expression of maintenance and de novo methyltransferase mRNA was measured using real-time RT-PCR. Analysis was performed on cDNA generated from total RNA from naïve, day 4, day 8, memory and exhausted LCMV-specific CD8 T cells. The difference in expression of isoform 1 and isoform 2 of the de novo methyltransferase Dnmt3a was determined using two different sets of primers that distinguish between the two isoforms. Only isoform 2 of Dnmt3a is differentially expressed between LCMV-specific CD8 T cells generated from acute and chronic infections. Transcript expression values were normalized to 18s ribosomal RNA.
Figure 7
PDCD1 CR-C is remethylated during effector to memory differentiation but remains demethylated in CD8 T cells specific to human viruses that cause chronic infections. A) Histogram analysis of PD-1 expression on naïve and virus-specific CD8 T cells. Bisulfite sequencing and graphical summary of CR-C of the PDCD1 promoter in human naïve (CCR7hi, CD45RAhi, PD-1lo), YF-17D-specific effector, and YF-17D-specific memory CD8 T cells. B) Histogram analysis of PD-1 expression on virus-specific CD8 T cells. Bisulfite sequencing and graphical summary of CR-C of the PDCD1 promoter in CMV and EBV-specific CD8 T cells (Cytomegalovirus & Epstein Barr virus). Class-I tetramers were used to sort antigen-specific cells from immune and chronically infected individuals. Cell purity post-sorting was ≥ 94%. Bisulfite sequencing and graphical summary of DNA from. Only CpG sites 18-26 are shown for clarity. CpG sites 1-17 reside inside the predicted CpG island which remains predominantly demethylated (Figure S5). Histogram analysis of PD-1 expression is above the corresponding methylation summary. PD-1 expression of the naïve cells is shown with the shaded gray line and PD-1 expression on the antigen-specific CD8 T cells is indicated by the open black line.
Comment in
- T cell memory: Exhausted T cells miss out on methylation.
Minton K. Minton K. Nat Rev Immunol. 2011 Oct 14;11(11):718-9. doi: 10.1038/nri3093. Nat Rev Immunol. 2011. PMID: 21997790 No abstract available.
Similar articles
- Demethylation of the PD-1 Promoter Is Imprinted during the Effector Phase of CD8 T Cell Exhaustion.
Ahn E, Youngblood B, Lee J, Lee J, Sarkar S, Ahmed R. Ahn E, et al. J Virol. 2016 Sep 12;90(19):8934-46. doi: 10.1128/JVI.00798-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27466420 Free PMC article. - Restoring function in exhausted CD8 T cells during chronic viral infection.
Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Barber DL, et al. Nature. 2006 Feb 9;439(7077):682-7. doi: 10.1038/nature04444. Epub 2005 Dec 28. Nature. 2006. PMID: 16382236 - Tissue-specific differences in PD-1 and PD-L1 expression during chronic viral infection: implications for CD8 T-cell exhaustion.
Blackburn SD, Crawford A, Shin H, Polley A, Freeman GJ, Wherry EJ. Blackburn SD, et al. J Virol. 2010 Feb;84(4):2078-89. doi: 10.1128/JVI.01579-09. Epub 2009 Dec 2. J Virol. 2010. PMID: 19955307 Free PMC article. - Costimulatory molecule programmed death-1 in the cytotoxic response during chronic hepatitis C.
Larrubia JR, Benito-Martínez S, Miquel J, Calvino M, Sanz-de-Villalobos E, Parra-Cid T. Larrubia JR, et al. World J Gastroenterol. 2009 Nov 7;15(41):5129-40. doi: 10.3748/wjg.15.5129. World J Gastroenterol. 2009. PMID: 19891011 Free PMC article. Review. - [Role of programmed death-1 in viral infectious diseases].
Lu FC, Nong GM. Lu FC, et al. Zhongguo Dang Dai Er Ke Za Zhi. 2018 Jan;20(1):77-82. doi: 10.7499/j.issn.1008-8830.2018.01.016. Zhongguo Dang Dai Er Ke Za Zhi. 2018. PMID: 29335088 Free PMC article. Review. Chinese.
Cited by
- Targeted Drug Screening Leveraging Senescence-Induced T-Cell Exhaustion Signatures in Hepatocellular Carcinoma.
Qi Q, Pang J, Chen Y, Tang Y, Wang H, Gul S, Sun Y, Tang W, Sheng M. Qi Q, et al. Int J Mol Sci. 2024 Oct 18;25(20):11232. doi: 10.3390/ijms252011232. Int J Mol Sci. 2024. PMID: 39457014 Free PMC article. - Epigenetic regulation of T cell adaptive immunity.
Frias AB, Boi SK, Lan X, Youngblood B. Frias AB, et al. Immunol Rev. 2021 Mar;300(1):9-21. doi: 10.1111/imr.12943. Epub 2021 Feb 28. Immunol Rev. 2021. PMID: 33644866 Free PMC article. Review. - Hypomethylation and up-regulation of PD-1 in T cells by azacytidine in MDS/AML patients: A rationale for combined targeting of PD-1 and DNA methylation.
Ørskov AD, Treppendahl MB, Skovbo A, Holm MS, Friis LS, Hokland M, Grønbæk K. Ørskov AD, et al. Oncotarget. 2015 Apr 20;6(11):9612-26. doi: 10.18632/oncotarget.3324. Oncotarget. 2015. PMID: 25823822 Free PMC article. - Generating long-lived CD8(+) T-cell memory: Insights from epigenetic programs.
Dogra P, Ghoneim HE, Abdelsamed HA, Youngblood B. Dogra P, et al. Eur J Immunol. 2016 Jul;46(7):1548-62. doi: 10.1002/eji.201545550. Eur J Immunol. 2016. PMID: 27230488 Free PMC article. Review. - The immunological synapse: the gateway to the HIV reservoir.
Kulpa DA, Brehm JH, Fromentin R, Cooper A, Cooper C, Ahlers J, Chomont N, Sékaly RP. Kulpa DA, et al. Immunol Rev. 2013 Jul;254(1):305-25. doi: 10.1111/imr.12080. Immunol Rev. 2013. PMID: 23772628 Free PMC article. Review.
References
- Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. - PubMed
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2 R37 AI30048-17/AI/NIAID NIH HHS/United States
- P01 AI080192/AI/NIAID NIH HHS/United States
- P01 AI080192-01/AI/NIAID NIH HHS/United States
- R37 AI030048-17/AI/NIAID NIH HHS/United States
- R37 AI030048/AI/NIAID NIH HHS/United States
- 1 P01 AI080192-01/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials