Identification of a functional transposon insertion in the maize domestication gene tb1 - PubMed (original) (raw)
Identification of a functional transposon insertion in the maize domestication gene tb1
Anthony Studer et al. Nat Genet. 2011.
Abstract
Genetic diversity created by transposable elements is an important source of functional variation upon which selection acts during evolution. Transposable elements are associated with adaptation to temperate climates in Drosophila, a SINE element is associated with the domestication of small dog breeds from the gray wolf and there is evidence that transposable elements were targets of selection during human evolution. Although the list of examples of transposable elements associated with host gene function continues to grow, proof that transposable elements are causative and not just correlated with functional variation is limited. Here we show that a transposable element (Hopscotch) inserted in a regulatory region of the maize domestication gene, teosinte branched1 (tb1), acts as an enhancer of gene expression and partially explains the increased apical dominance in maize compared to its progenitor, teosinte. Molecular dating indicates that the Hopscotch insertion predates maize domestication by at least 10,000 years, indicating that selection acted on standing variation rather than new mutation.
Figures
Figure 1
Teosinte and maize plants. (a) Highly branched teosinte plant. (b) Teosinte lateral branch with terminal tassel. (c) Unbranched maize plant. (d) Maize ear shoot (that is, lateral branch).
Figure 2
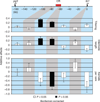
The phenotypic additive effects for seven intervals across the tb1 genomic region. The horizontal axis represents the tb1 genomic region to scale. Base-pair positions are relative to AGPv2 position 265,745,977 of the maize reference genome sequence. The tb1 ORF and the nearest upstream predicted gene (pg3) are shown. The previously defined control region (CR) is shown in red and is divided into its proximal and distal components. Vertical columns represent the additive effects shown with standard error bars for each of the three traits in each of the seven intervals that were tested for an effect on phenotype. Black columns are statistically significant (P (Bonferroni) < 0.05); white bars are not statistically significant (_P_ (Bonferroni) > 0.05).
Figure 3
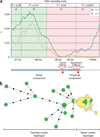
Sequence diversity in maize and teosinte across the control region. (a) Nucleotide diversity across the tb1 upstream control region. Base-pair positions are relative to AGPv2 position 265,745,977 of the maize reference genome sequence. P values correspond to HKA neutrality tests for regions A–D, as defined by the dotted lines. Green shading signifies evidence of neutrality, and pink shading signifies regions of non-neutral evolution. Nucleotide diversity (π) for maize (yellow line) and teosinte (green line) were calculated using a 500-bp sliding window with a 25-bp step. The distal and proximal components of the control region with four fixed sequence differences between the most common maize haplotype and teosinte haplotype are shown below. (b) A minimum spanning tree for the control region with 16 diverse maize and 17 diverse teosinte sequences. Size of the circles for each haplotype group (yellow, maize; green, teosinte) is proportional to the number of individuals within that haplotype.
Figure 4
Constructs and corresponding normalized luciferase expression levels. Transient assays were performed in maize leaf protoplast. Each construct is drawn to scale. The construct backbone consists of the minimal promoter from the cauliflower mosaic virus (mpCaMV, gray box), luciferase ORF (luc, white box) and the nopaline synthase terminator (black box). Portions of the proximal and distal components of the control region (hatched boxes) from maize and teosinte were cloned into restriction sites upstream of the minimal promoter. “Δ” denotes the excision of either the Tourist or Hopscotch element from the maize construct. Horizontal green bars show the normalized mean with s.e.m. for each construct.
Comment in
- A transposon in tb1 drove maize domestication.
Tsiantis M. Tsiantis M. Nat Genet. 2011 Oct 27;43(11):1048-50. doi: 10.1038/ng.986. Nat Genet. 2011. PMID: 22030605 No abstract available.
Similar articles
- Natural variation in teosinte at the domestication locus teosinte branched1 (tb1).
Vann L, Kono T, Pyhäjärvi T, Hufford MB, Ross-Ibarra J. Vann L, et al. PeerJ. 2015 Apr 16;3:e900. doi: 10.7717/peerj.900. eCollection 2015. PeerJ. 2015. PMID: 25909039 Free PMC article. - The evolution of apical dominance in maize.
Doebley J, Stec A, Hubbard L. Doebley J, et al. Nature. 1997 Apr 3;386(6624):485-8. doi: 10.1038/386485a0. Nature. 1997. PMID: 9087405 - Estimating a nucleotide substitution rate for maize from polymorphism at a major domestication locus.
Clark RM, Tavaré S, Doebley J. Clark RM, et al. Mol Biol Evol. 2005 Nov;22(11):2304-12. doi: 10.1093/molbev/msi228. Epub 2005 Aug 3. Mol Biol Evol. 2005. PMID: 16079248 - Maize domestication and gene interaction.
Stitzer MC, Ross-Ibarra J. Stitzer MC, et al. New Phytol. 2018 Oct;220(2):395-408. doi: 10.1111/nph.15350. Epub 2018 Jul 23. New Phytol. 2018. PMID: 30035321 Review. - Genomic screening for artificial selection during domestication and improvement in maize.
Yamasaki M, Wright SI, McMullen MD. Yamasaki M, et al. Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review.
Cited by
- Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture.
Baumann K, Venail J, Berbel A, Domenech MJ, Money T, Conti L, Hanzawa Y, Madueno F, Bradley D. Baumann K, et al. J Exp Bot. 2015 Aug;66(15):4769-80. doi: 10.1093/jxb/erv247. Epub 2015 May 27. J Exp Bot. 2015. PMID: 26019254 Free PMC article. - Unraveling the KNOTTED1 regulatory network in maize meristems.
Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O'Connor D, Grotewold E, Hake S. Bolduc N, et al. Genes Dev. 2012 Aug 1;26(15):1685-90. doi: 10.1101/gad.193433.112. Genes Dev. 2012. PMID: 22855831 Free PMC article. - European flint landraces grown in situ reveal adaptive introgression from modern maize.
Bitocchi E, Bellucci E, Rau D, Albertini E, Rodriguez M, Veronesi F, Attene G, Nanni L. Bitocchi E, et al. PLoS One. 2015 Apr 8;10(4):e0121381. doi: 10.1371/journal.pone.0121381. eCollection 2015. PLoS One. 2015. PMID: 25853809 Free PMC article. - The contribution of transposable elements to transcriptional novelty in plants: the FLC affair.
Quadrana L. Quadrana L. Transcription. 2020 Jun-Aug;11(3-4):192-198. doi: 10.1080/21541264.2020.1803031. Epub 2020 Aug 12. Transcription. 2020. PMID: 32783496 Free PMC article. Review. - The impact of transposable elements on tomato diversity.
Domínguez M, Dugas E, Benchouaia M, Leduque B, Jiménez-Gómez JM, Colot V, Quadrana L. Domínguez M, et al. Nat Commun. 2020 Aug 13;11(1):4058. doi: 10.1038/s41467-020-17874-2. Nat Commun. 2020. PMID: 32792480 Free PMC article.
References
- Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. - PubMed
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. - PubMed
- Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retrotransposon. Nature. 2006;441:87–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources