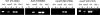The iron-repressed, AraC-like regulator MpeR activates expression of fetA in Neisseria gonorrhoeae - PubMed (original) (raw)
The iron-repressed, AraC-like regulator MpeR activates expression of fetA in Neisseria gonorrhoeae
Aimee Hollander et al. Infect Immun. 2011 Dec.
Abstract
Neisseria gonorrhoeae is an obligate human pathogen that causes the common sexually transmitted infection gonorrhea. Gonococcal infections cause significant morbidity, particularly among women, as the organism ascends to the upper reproductive tract, resulting in pelvic inflammatory disease, ectopic pregnancy, and infertility. In the last few years, antibiotic resistance rates have risen dramatically, leading to severe restriction of treatment options for gonococcal disease. Gonococcal infections do not elicit protective immunity, nor is there an effective vaccine to prevent the disease. Thus, further understanding of the expression, function, and regulation of surface antigens could lead to better treatment and prevention modalities in the future. In the current study, we determined that an iron-repressed regulator, MpeR, interacted specifically with the DNA sequence upstream of fetA and activated FetA expression. Interestingly, MpeR was previously shown to regulate the expression of gonococcal antimicrobial efflux systems. We confirmed that the outer membrane transporter FetA allows gonococcal strain FA1090 to utilize the xenosiderophore ferric enterobactin as an iron source. However, we further demonstrated that FetA has an extended range of substrates that encompasses other catecholate xenosiderophores, including ferric salmochelin and the dimers and trimers of dihydroxybenzoylserine. We demonstrated that fetA is part of an iron-repressed, MpeR-activated operon which putatively encodes other iron transport proteins. This is the first study to describe a regulatory linkage between antimicrobial efflux and iron transport in N. gonorrhoeae. The regulatory nidus that links these systems, MpeR, is expressed exclusively by pathogenic neisseriae and is therefore expected to be an important virulence factor.
Figures
Fig. 1.
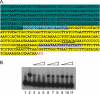
FetA expression is regulated by MpeR. (A) SDS-PAGE analysis of protein expression. Total membrane proteins were isolated from wild-type (WT) (FA1090) and mpeR mutant (MCV304) strains grown under iron-depleted (−) and iron-replete (+) conditions for 4 h. Proteins were separated on a 7.5% acrylamide gel. The arrow on the left indicates the band that was excised and identified as FetA by mass spectrometry analysis. The positions of molecular mass markers are indicated on the right. (B) Western blot analysis of FetA expression. The WT (FA1090), mpeR mutant (MCV304), fetA mutant (FA6959), and complemented _mpeR_C (MCV305) strains were grown under iron-depleted (−) and iron-replete (+) conditions. Total membrane proteins from each strain were isolated and standardized before being separated by SDS-PAGE and then transferred to nitrocellulose. Blots were probed with an anti-FetA monoclonal antibody.
Fig. 2.
MpeR activates fetA transcription under iron-depleted conditions. The WT (FA1090), fetA mutant (FA6959), mpeR mutant (MCV304), and complemented _mpeR_C (MCV305) strains were grown under iron-depleted (−) and iron-replete (+) conditions. RNA samples isolated from each gonococcal strain were analyzed for expression of fetA and mpeR by RT-PCR. The 16S rRNA gene (16s)was used as a positive control because it was constitutively expressed under all growth conditions. Expression of the 16S rRNA gene in the absence of reverse transcriptase [16s(−)] was used as a negative control.
Fig. 3.
MpeR binds upstream of fetA in a specific manner. (A) Sequence of the 500-bp intergenic region immediately upstream of fetA. The sequence highlighted in teal is contained within the _fetA_1 probe employed for the EMSA shown in panel B. The sequence highlighted in yellow is contained within the _fetA_2 competitor DNA. The sequence highlighted in blue is contained in both _fet_A1 and _fetA_2 amplicons. The Fur binding site (34) is highlighted in gray. The previously mapped (14) promoter elements (underlined) and transcriptional start site (asterisk) are identified. The start codon for FetA is shown in red. (B) Five nanograms of the 250-bp labeled _fetA_1 probe (lane 1) was incubated with 10 μg of MBP-MpeR in the absence of unlabeled competitor (lane 2) or in the presence of 2×, 10×, or 20× excess unlabeled competitor DNA. Lanes 3 to 5 contain reaction mixtures including increasing concentrations of the specific competitor (_fetA_1), lanes 6 to 8 contain mixtures including increasing concentrations of _fetA_2, and lanes 9 to 11 contain mixtures including increasing concentrations of the nonspecific competitor rnpB.
Fig. 4.
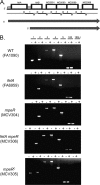
FetA is encoded as part of a multigene operon. (A) Genetic locus, including fetA and downstream genes. Genes are depicted by boxes. Hatched regions 5′ of fetA and fetB indicate approximate locations of Fur boxes (34). Below the chromosomal locus are small dark arrows indicating primer locations. Numbered black bars denote the amplicons generated from each primer set. Long, dark gray arrows indicate the lengths and start positions of two proposed transcripts. (B) RT-PCR analysis of the fet operon. RNA was isolated from the indicated gonococcal strains which were grown under iron-depleted (−) and iron-replete (+) conditions. Amplicon numbers correspond to the diagram in panel A. The 16S rRNA gene (16S) was used as a positive control because it is constitutively expressed under all conditions tested. Expression of the 16S rRNA gene in the absence of reverse transcriptase [16S(−)] was used as a negative control.
Fig. 5.
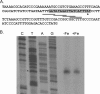
Identification of the fetB transcriptional start site. (A) Sequence of the intergenic region immediately upstream of fetB. The ATG at the end of the sequence represents the FetB start codon. Several overlapping potential −10 promoter elements are underlined. The Fur binding site (34) is highlighted in gray. The transcriptional start site identified in this analysis is identified by the asterisk. (B) Primer extension products generated from RNA samples harvested from wild-type gonococcal strain FA1090 grown under iron-replete (+Fe) and iron-depleted (−Fe) conditions. Equivalency of the amount of RNA template in each sample was confirmed by ethidium bromide staining of RNA separated on an agarose gel. For comparison, the sequencing reaction using the same primer as was used for the primer extension reaction is shown on the left. The T residue highlighted by the asterisk marks the point of transcript initiation on the noncoding strand.
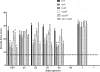
Fig. 6.
Xenosiderophore utilization by gonococcal strain FA1090. CDM plates were supplemented with apo-bovine transferrin, and wells within the plates were inoculated with the following siderophores: ENT, enterobactin; D1, dihydroxybenzoylserine (DHBS); D2, the dimer form of DHBS; D3, the trimer form of DHBS; S2, the linear derivative of salmochelin; and S4, the cyclized form of salmochelin. Ferric citrate (+) was used as the positive control, and apo-bovine transferrin (−) was used as the negative control. Each bar indicates the average growth in millimeters around each siderophore source; the averages and standard deviations were determined from seven independent experiments, each conducted in triplicate. Bars represent average growth zones for the wild-type strain FA1090, fetA mutant strain FA6959, tonB mutant strain MCV656, the mpeR mutant strain, the _mpeR_C complemented strain, and the fetA mpeR double mutant strain. The horizontal dashed line indicates the diameter of the well containing each iron source. Pairwise comparisons between the wild-type and mutant strains resulted in the following P values: *, P < 0.001; #, P = 0.0124; ^, P = 0.0197.
Fig. 7.
FetA expression is not induced by the presence of xenosiderophores. (A) WT strain FA1090 was grown in CDM with the indicated ferrated xenosiderophores (final concentration of 10 μM) as the sole iron source: ENT, enterobactin; D1, dihydroxybenzoylserine (DHBS); D2, the dimer form of DHBS; D3, the trimer form of DHBS; S2, the linear derivative of salmochelin; and S4, the cyclized form of salmochelin. As controls, the WT strain was grown in the absence of iron (−) or with ferric nitrate (+) but without the addition of siderophores. Aliquots collected at 2, 4, and 6 h (indicated above the blots) were lysed and subjected to SDS-PAGE. After separation, proteins were transferred to nitrocellulose. Blots were probed with anti-FetA (top) or anti-TbpA (bottom) antibodies. (B) As in panel A, except the WT strain FA1090 was grown in CDM with the indicated xenosiderophores in the iron-free or apo form.
Similar articles
- The fbpABC operon is required for Ton-independent utilization of xenosiderophores by Neisseria gonorrhoeae strain FA19.
Strange HR, Zola TA, Cornelissen CN. Strange HR, et al. Infect Immun. 2011 Jan;79(1):267-78. doi: 10.1128/IAI.00807-10. Epub 2010 Nov 1. Infect Immun. 2011. PMID: 21041493 Free PMC article. - Ferric enterobactin binding and utilization by Neisseria gonorrhoeae.
Carson SD, Klebba PE, Newton SM, Sparling PF. Carson SD, et al. J Bacteriol. 1999 May;181(9):2895-901. doi: 10.1128/JB.181.9.2895-2901.1999. J Bacteriol. 1999. PMID: 10217784 Free PMC article. - MpeR regulates the mtr efflux locus in Neisseria gonorrhoeae and modulates antimicrobial resistance by an iron-responsive mechanism.
Mercante AD, Jackson L, Johnson PJ, Stringer VA, Dyer DW, Shafer WM. Mercante AD, et al. Antimicrob Agents Chemother. 2012 Mar;56(3):1491-501. doi: 10.1128/AAC.06112-11. Epub 2012 Jan 3. Antimicrob Agents Chemother. 2012. PMID: 22214775 Free PMC article. - Transcriptional regulation of the mtrCDE efflux pump operon: importance for Neisseria gonorrhoeae antimicrobial resistance.
Ayala JC, Balthazar JT, Shafer WM. Ayala JC, et al. Microbiology (Reading). 2022 Aug;168(8):001231. doi: 10.1099/mic.0.001231. Microbiology (Reading). 2022. PMID: 35916832 Free PMC article. Review. - The ironclad truth: how in vivo transcriptomics and in vitro mechanistic studies shape our understanding of Neisseria gonorrhoeae gene regulation during mucosal infection.
Moreau MR, Massari P, Genco CA. Moreau MR, et al. Pathog Dis. 2017 Jul 31;75(5):ftx057. doi: 10.1093/femspd/ftx057. Pathog Dis. 2017. PMID: 28520925 Free PMC article. Review.
Cited by
- Stealthy microbes: How Neisseria gonorrhoeae hijacks bulwarked iron during infection.
Stoudenmire JL, Greenawalt AN, Cornelissen CN. Stoudenmire JL, et al. Front Cell Infect Microbiol. 2022 Sep 15;12:1017348. doi: 10.3389/fcimb.2022.1017348. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36189345 Free PMC article. Review. - Molecular Regulatory Mechanisms Drive Emergent Pathogenetic Properties of Neisseria gonorrhoeae.
Sunkavalli A, McClure R, Genco C. Sunkavalli A, et al. Microorganisms. 2022 Apr 28;10(5):922. doi: 10.3390/microorganisms10050922. Microorganisms. 2022. PMID: 35630366 Free PMC article. Review. - Viral vectors expressing group B meningococcal outer membrane proteins induce strong antibody responses but fail to induce functional bactericidal activity.
Marsay L, Dold C, Paterson GK, Yamaguchi Y, Derrick JP, Chan H, Feavers IM, Maiden MCJ, Wyllie D, Hill AV, Pollard AJ, Rollier CS. Marsay L, et al. J Infect. 2022 May;84(5):658-667. doi: 10.1016/j.jinf.2022.02.032. Epub 2022 Mar 1. J Infect. 2022. PMID: 35245584 Free PMC article. - Human B Cell Responses to Dominant and Subdominant Antigens Induced by a Meningococcal Outer Membrane Vesicle Vaccine in a Phase I Trial.
Rollier CS, Dold C, Marsay L, Linder A, Green CA, Sadarangani M, Norheim G, Derrick JP, Feavers IM, Maiden MCJ, Pollard AJ. Rollier CS, et al. mSphere. 2022 Feb 23;7(1):e0067421. doi: 10.1128/msphere.00674-21. Epub 2022 Jan 26. mSphere. 2022. PMID: 35080470 Free PMC article. Clinical Trial. - The Role of the Moraxella catarrhalis CopB Protein in Facilitating Iron Acquisition From Human Transferrin and Lactoferrin.
Chan C, Ng D, Schryvers AB. Chan C, et al. Front Microbiol. 2021 Sep 23;12:714815. doi: 10.3389/fmicb.2021.714815. eCollection 2021. Front Microbiol. 2021. PMID: 34630348 Free PMC article.
References
- Ala'Aldeen D. A., Wall R. A., Borriello S. P. 1990. Immunogenicity and cross-reactivity of the 70-Kda iron-regulated protein of Neisseria meningitidis in man and animals. J. Med. Microbiol. 32: 275–281 - PubMed
- Anderson J. E., Hobbs M. M., Biswas G. D., Sparling P. F. 2003. Opposing selective forces for expression of the gonococcal lactoferrin receptor. Mol. Microbiol. 48: 1325–1337 - PubMed
- Archibald F. S., Simonson C., DeVoe I. W. 1981. Comparison of iron binding and uptake from FeCl3 and Fe-citrated by Neisseria meningitidis. Can. J. Microbiol. 27: 1066–1070 - PubMed
- Bagg A., Neilands J. B. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26: 5471–5477 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI065555/AI/NIAID NIH HHS/United States
- U19 AI031496/AI/NIAID NIH HHS/United States
- AI065555/AI/NIAID NIH HHS/United States
- R01 AI047141/AI/NIAID NIH HHS/United States
- AI084400/AI/NIAID NIH HHS/United States
- R01 AI084400/AI/NIAID NIH HHS/United States
- R37 AI021150/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical