Proteasome functional insufficiency in cardiac pathogenesis - PubMed (original) (raw)
Review
Proteasome functional insufficiency in cardiac pathogenesis
Xuejun Wang et al. Am J Physiol Heart Circ Physiol. 2011 Dec.
Abstract
The ubiquitin-proteasome system (UPS) is responsible for the degradation of most cellular proteins. Alterations in cardiac UPS, including changes in the degradation of regulatory proteins and proteasome functional insufficiency, are observed in many forms of heart disease and have been shown to play an important role in cardiac pathogenesis. In the past several years, remarkable progress in understanding the mechanisms that regulate UPS-mediated protein degradation has been achieved. A transgenic mouse model of benign enhancement of cardiac proteasome proteolytic function has been created. This has led to the first demonstration of the necessity of proteasome functional insufficiency in the genesis of important pathological processes. Cardiomyocyte-restricted enhancement of proteasome proteolytic function by overexpression of proteasome activator 28α protects against cardiac proteinopathy and myocardial ischemia-reperfusion injury. Additionally, exciting advances have recently been achieved in the search for a pharmacological agent to activate the proteasome. These breakthroughs are expected to serve as an impetus to further investigation into the involvement of UPS dysfunction in molecular pathogenesis and to the development of new therapeutic strategies for combating heart disease. An interplay between the UPS and macroautophagy is increasingly suggested in noncardiac systems but is not well understood in the cardiac system. Further investigations into the interplay are expected to provide a more comprehensive picture of cardiac protein quality control and degradation.
Figures
Fig. 1.
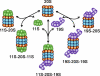
A transgenic mouse model expressing carboxyl fusion of degron CL1 to green fluorescent protein (GFPdgn) as a surrogate misfolded protein. A: an illustration of the design of GFPdgn transgene and the sequence of degron CL1. GFPdgn is a fusion protein derived from carboxyl fusion of an enhanced GFP with degron CL1, which contains the depicted 16 amino acid residues. B: sequence and helical wheel representation of degron CL1 [adopted from Gilon et al. with permission (37)]. The hydrophobic amino acid residues are boxed. Note that in its helical structure, degron CL1 contains surface exposure of a long patch of hydrophobic amino acid residues. C: representative images of Western blot analysis of the indicated proteins in cardiac and hepatic tissues from adult GFPdgn transgenic mice (line 3) 20 h after an intraperitoneal injection of proteasome inhibitor MG-262 (5 μmol/kg) or the vehicle control. At the baseline, GFPdgn is expressed at a higher level in the heart than in the liver. MG-262 treatment increases ubiquitinated proteins, the precursor of the β5-subunit of the 20S proteasome (pre-β5) and GFPdgn proteins in both organs shown here. Ub, ubiquitin; NS, not significant.
Fig. 2.
A schematic depiction of proteasome complexes expressed in mammalian cells. The 20S proteasome harbors all the proteolytic activities of the proteasome. To degrade proteins, the 20S needs to team up with the 19S and/or the 11S proteasome activator.
Fig. 3.
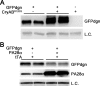
Use of GFPdgn reporter mice reveals proteasome functional insufficiency (PFI, A) and proteasome functional enhancement (B) in the heart. A: representative image of Western blot analyses for GFPdgn in the GFPdgn/CryABR120G double transgenic mouse heart, compared with littermate GFPdgn single transgenic mouse hearts. The soluble fraction of myocardial protein extracts from 1-mo-old mice of the indicated genotypes (top labels) were used for the analyses. PFI in the CryABR120G-based desmin-related cardiomyopathy mouse heart is revealed by the increased GFPdgn protein levels in the double transgenic mouse hearts. B: representative image of Western blot analyses for GFPdgn and proteasome activator 28α (PA28α) in the heart. Total proteins were extracted from ventricular myocardium of littermate mice of the indicated genotypes (labels at the top of the panel) at 8 wk of age and used for Western blot analyses for the indicated proteins. Note that GFPdgn protein levels were markedly decreased in PA28α overexpression hearts, indicating that proteasome proteolytic function in the heart is significantly enhanced by cardiomyocyte-restricted overexpression of PA28α. LC, loading control; tTA, tetracycline-controlled transactivator.
Fig. 4.
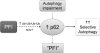
An illustration of a working hypothesis on the interplay between proteasome-mediated proteolysis and the selective autophagy. On the one hand, PFI accumulates ubiquitin conjugates which, through interaction, retain p62 proteins in the cell. PFI may also activate the nuclear factor erythroid-derived 2-related factor-1 (Nrf2)-mediated transcriptional program to activate p62 synthesis. The binding of p62 to ubiquitin conjugates in protein aggregates may recruit autophagosomes to and remove the aggregates, thereby increasing selective autophagy. On the other hand, chronic autophagy inhibition/impairment can accumulate p62, which is a substrate of autophagy. The accumulated p62 can bind to ubiquitinated proteins formed during normal ubiquitination and hinder them from being delivered to the proteasome for degradation; therefore, autophagy impairment may indirectly induce a PFI-like phenotype, although proteasome assembly and activities may not be decreased.
Similar articles
- The role of the proteasome in heart disease.
Li YF, Wang X. Li YF, et al. Biochim Biophys Acta. 2011 Feb;1809(2):141-9. doi: 10.1016/j.bbagrm.2010.09.001. Epub 2010 Sep 15. Biochim Biophys Acta. 2011. PMID: 20840877 Free PMC article. Review. - Inadequate ubiquitination-proteasome coupling contributes to myocardial ischemia-reperfusion injury.
Hu C, Tian Y, Xu H, Pan B, Terpstra EM, Wu P, Wang H, Li F, Liu J, Wang X. Hu C, et al. J Clin Invest. 2018 Dec 3;128(12):5294-5306. doi: 10.1172/JCI98287. Epub 2018 Oct 22. J Clin Invest. 2018. PMID: 30204128 Free PMC article. - p62 Stages an interplay between the ubiquitin-proteasome system and autophagy in the heart of defense against proteotoxic stress.
Su H, Wang X. Su H, et al. Trends Cardiovasc Med. 2011 Nov;21(8):224-8. doi: 10.1016/j.tcm.2012.05.015. Trends Cardiovasc Med. 2011. PMID: 22902070 Free PMC article. Review. - Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice.
Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Li J, et al. J Clin Invest. 2011 Sep;121(9):3689-700. doi: 10.1172/JCI45709. Epub 2011 Aug 15. J Clin Invest. 2011. PMID: 21841311 Free PMC article. - Targeting the ubiquitin-proteasome system in heart disease: the basis for new therapeutic strategies.
Drews O, Taegtmeyer H. Drews O, et al. Antioxid Redox Signal. 2014 Dec 10;21(17):2322-43. doi: 10.1089/ars.2013.5823. Epub 2014 Oct 1. Antioxid Redox Signal. 2014. PMID: 25133688 Free PMC article. Review.
Cited by
- Aging in Heart Failure: Embracing Biology Over Chronology: JACC Family Series.
Goyal P, Maurer MS, Roh J. Goyal P, et al. JACC Heart Fail. 2024 May;12(5):795-809. doi: 10.1016/j.jchf.2024.02.021. Epub 2024 Apr 8. JACC Heart Fail. 2024. PMID: 38597865 Free PMC article. Review. - Genetic ablation of Lmp2 increases the susceptibility for impaired cardiac function.
Trogisch FA, Koser F, Michel S, Liem DA, Florea BI, Hecker M, Drews O. Trogisch FA, et al. Front Mol Biosci. 2024 Mar 7;11:1148948. doi: 10.3389/fmolb.2024.1148948. eCollection 2024. Front Mol Biosci. 2024. PMID: 38516190 Free PMC article. - Zonisamide attenuates pressure overload-induced myocardial hypertrophy in mice through proteasome inhibition.
Wu Q, Liu WJ, Ma XY, Chang JS, Zhao XY, Liu YH, Yu XY. Wu Q, et al. Acta Pharmacol Sin. 2024 Apr;45(4):738-750. doi: 10.1038/s41401-023-01191-7. Epub 2023 Dec 14. Acta Pharmacol Sin. 2024. PMID: 38097716 - S14-Phosphorylated RPN6 Mediates Proteasome Activation by PKA and Alleviates Proteinopathy.
Yang L, Parajuli N, Wu P, Liu J, Wang X. Yang L, et al. Circ Res. 2023 Sep 15;133(7):572-587. doi: 10.1161/CIRCRESAHA.123.322887. Epub 2023 Aug 29. Circ Res. 2023. PMID: 37641975 Free PMC article. - Protein Quality Control at the Sarcomere: Titin Protection and Turnover and Implications for Disease Development.
Kötter S, Krüger M. Kötter S, et al. Front Physiol. 2022 Jun 30;13:914296. doi: 10.3389/fphys.2022.914296. eCollection 2022. Front Physiol. 2022. PMID: 35846001 Free PMC article. Review.
References
- Adams V, Linke A, Wisloff U, Doring C, Erbs S, Krankel N, Witt CC, Labeit S, Muller-Werdan U, Schuler G, Hambrecht R. Myocardial expression of Murf-1 and MAFbx after induction of chronic heart failure: effect on myocardial contractility. Cardiovasc Res 73: 120–129, 2007 - PubMed
- Ahmad K. Proteasome inhibitor for treatment of multiple myeloma. Lancet Oncol 6: 546, 2005 - PubMed
- Amici M, Lupidi G, Angeletti M, Fioretti E, Eleuteri AM. Peroxynitrite-induced oxidation and its effects on isolated proteasomal systems. Free Radic Biol Med 34: 987–996, 2003 - PubMed
- Asai A, Tanahashi N, Qiu JH, Saito N, Chi S, Kawahara N, Tanaka K, Kirino T. Selective proteasomal dysfunction in the hippocampal CA1 region after transient forebrain ischemia. J Cereb Blood Flow Metab 22: 705–710, 2002 - PubMed
- Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, Takahama H, Sasaki H, Higo S, Asakura M, Takashima S, Hori M, Kitakaze M. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. J Mol Cell Cardiol 46: 452–462, 2009 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL-0668936/HL/NHLBI NIH HHS/United States
- HL-085629/HL/NHLBI NIH HHS/United States
- HL-072166/HL/NHLBI NIH HHS/United States
- R01 HL085629/HL/NHLBI NIH HHS/United States
- R01 HL072166/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous