Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington's disease - PubMed (original) (raw)
Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington's disease
Ryan L Boudreau et al. Mol Ther. 2011 Dec.
Abstract
RNA interference (RNAi) provides an approach for the treatment of many human diseases. However, the safety of RNAi-based therapies can be hampered by the ability of small inhibitory RNAs (siRNAs) to bind to unintended mRNAs and reduce their expression, an effect known as off-target gene silencing. Off-targeting primarily occurs when the seed region (nucleotides 2-8 of the small RNA) pairs with sequences in 3'-UTRs of unintended mRNAs and directs translational repression and destabilization of those transcripts. To date, most therapeutic RNAi sequences are selected primarily for gene silencing efficacy, and later evaluated for safety. Here, in designing siRNAs to treat Huntington's disease (HD), a dominant neurodegenerative disorder, we prioritized selection of sequences with minimal off-targeting potentials (i.e., those with a scarcity of seed complements within all known human 3'-UTRs). We identified new promising therapeutic candidate sequences which show potent silencing in cell culture and mouse brain. Furthermore, we present microarray data demonstrating that off-targeting is significantly minimized by using siRNAs that contain "safe" seeds, an important strategy to consider during preclinical development of RNAi-based therapeutics.
Figures
Figure 1
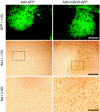
The artificial microRNA (miRNA), miSCR, causes neurotoxicity in mouse brain. Wild-type mice were injected into the striatum with adeno-associated virus (AAV)-green fluorescent protein (GFP) (expresses GFP only) or AAV-miSCR-GFP (expresses both the artificial miRNA and GFP), and histological analyses were performed on brains harvested at 6 months post-treatment. Photomicrographs representing GFP autofluorescence and immunohistochemical staining of IbaI-positive microglia are shown. Bars = 200 and 50 µm for ×10 and ×40 images, respectively. Boxed inset represents region of ×40 image.
Figure 2
Overview of seed-related off-targeting: mechanism and probabilities. (a) Diagram depicting the expression and processing of an artificial microRNA (miRNA) to produce the mature small inhibitory RNA (siRNA) duplex. The antisense guide strand is loaded into RNA-induced silencing complex (RISC) and may direct on-target silencing (intended) and off-target silencing (unintended). (b) Cartoon highlighting the relationship between the frequencies of seed complement binding sites in the 3′-UTRome and the off-targeting potential for siRNAs. (c) The number of human mRNA 3′-UTRs containing a given hexamer was determined for all of the 4,096 possible hexamers and a binned distribution is shown. The probabilities that randomly selected siRNAs targeting human coding sequence (CDS) will contain seed complements in a given range (white and gray shading) are also presented. For example, there is only a 10% chance that a randomly selected siRNA contains a seed complement for a hexamer present in ~1,500 human 3′-UTRs or less. Note: the sequences tested in this article are placed above their respective ranges.
Figure 3
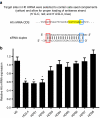
Selection and screening of huntingtin (htt)-targeting small inhibitory RNAs (siRNAs) with low off-targeting potentials. (a) Schematic outlining the selection of “safe” seed siRNAs with proper strand-biasing. (b) Plasmids expressing artificial microRNAs (miRNAs), harboring the indicated siRNA sequences, were transfected into HEK293 cells, and quantitative PCR (QPCR) analysis was performed 24 hours later to measure endogenous htt mRNA levels. U6 (promoter-only) and HD2.4 (a previously published htt RNAi sequence) serve as the negative and positive controls, respectively. Results are shown as mean ± SEM (N = 6, *P < 0.001, relative to U6).
Figure 4
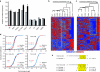
Evaluation of microarray data for huntingtin (htt) silencing and off-targeting. HEK293 cells were transfected with U6 promoter-only or U6-driven artificial microRNA (miRNA) expression plasmids (n = 4 for each treatment), and RNA was harvested 72 hours later for microarray analysis. Two-way ANOVA was performed to detect differentially expressed genes among the treatment groups. (a) Htt mRNA levels determined by microarray (gray bars) were consistent with those measured by quantitative PCR (QPCR) (black bars) using the same RNA samples. (b) Hierarchical clustering and heat-maps were generated using differentially expressed genes (P < 0.0001, 827 genes) to visualize the relationships among the treatment groups. Interestingly, all of the “safe” seed sequences are more related to U6 than the remaining sequences predicted to have higher off-targeting potentials (boundary marked by white line). (**c**) Hierarchical clustering and heat-maps were generated using differentially expressed genes (_P_ < 0.01, 985 genes) to visualize the relationships among the treatment groups. The impact of seed sequence on gene expression can be appreciated by the clustering of HD8.2 and 8.2mis which share the same seed. Notably, the predicted low off-targeting sequences (Safe, HDS1 and HDS2) are more similar to U6 and have smaller off-targeting signatures compared to both HD2.4 and HD8.2. Seed-related off-targeting was evaluated by (**d**) cumulative distribution and (**e**) motif discovery analyses. (**d**) Cumulative distribution plots for gene expression values are shown for transcripts containing (1 site or 2+ sites) or lacking (baseline) 3′-UTR seed complement binding sites for the indicated sequence and strand. A shift to the left indicates an increased likelihood of being downregulated. AS, antisense, S, sense. Kolmogorov–Smirnov (KS)-test _P_ values are shown; N.S., no statistical significance (_P_ > 0.1). (e) Motif discovery analyses identified an enrichment of seed complement binding sites in the 3′-UTRs of downregulated genes (>1.1-fold) unique to each treatment. Shown here are the examples of 8.2-124a and Terror; similar data for the remaining sequences supports that each mediates detectable seed-related off-targeting to some degree (Supplementary Figure S1).
Figure 5
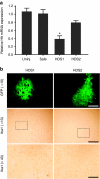
Silencing efficacy and safety of HDS sequences in mouse brain. Wild-type mice were injected into the striatum with adeno-associated virus (AAV) viruses coexpressing artificial microRNAs (miRNAs) and green fluorescent protein (GFP). (a) At 3 weeks postinjection, GFP-positive striata were harvested and quantitative PCR (QPCR) analysis was performed to measure endogenous mouse huntingtin (htt) mRNA levels. Results are shown as mean ± SEM (n ≥ 3, *P = 0.001, relative to uninjected striata). (b) Brains from additional cohorts of injected mice were harvested at 6 months postinjection and histological analyses were performed to assess neurotoxicity. Photomicrographs representing GFP autofluorescence and immunohistochemical staining of IbaI-positive microglia are shown. Bars = 200 and 50 µm for ×10 and ×40 images, respectively. Boxed inset represents region of ×40 image.
Similar articles
- Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease.
McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR, Davidson BL. McBride JL, et al. Mol Ther. 2011 Dec;19(12):2152-62. doi: 10.1038/mt.2011.219. Epub 2011 Oct 25. Mol Ther. 2011. PMID: 22031240 Free PMC article. - Chemical engineering of therapeutic siRNAs for allele-specific gene silencing in Huntington's disease models.
Conroy F, Miller R, Alterman JF, Hassler MR, Echeverria D, Godinho BMDC, Knox EG, Sapp E, Sousa J, Yamada K, Mahmood F, Boudi A, Kegel-Gleason K, DiFiglia M, Aronin N, Khvorova A, Pfister EL. Conroy F, et al. Nat Commun. 2022 Oct 3;13(1):5802. doi: 10.1038/s41467-022-33061-x. Nat Commun. 2022. PMID: 36192390 Free PMC article. - Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi.
McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. McBride JL, et al. Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5868-73. doi: 10.1073/pnas.0801775105. Epub 2008 Apr 8. Proc Natl Acad Sci U S A. 2008. PMID: 18398004 Free PMC article. - Nucleic Acid Therapeutics in Huntington's Disease.
Singh K, Roy I. Singh K, et al. Recent Pat Biotechnol. 2019;13(3):187-206. doi: 10.2174/1872208313666190208163714. Recent Pat Biotechnol. 2019. PMID: 30747088 Review. - Short interfering RNA (siRNA) as a novel therapeutic.
Pushparaj PN, Melendez AJ. Pushparaj PN, et al. Clin Exp Pharmacol Physiol. 2006 May-Jun;33(5-6):504-10. doi: 10.1111/j.1440-1681.2006.04399.x. Clin Exp Pharmacol Physiol. 2006. PMID: 16700886 Retracted. Review.
Cited by
- Recombinant AAV as a platform for translating the therapeutic potential of RNA interference.
Borel F, Kay MA, Mueller C. Borel F, et al. Mol Ther. 2014 Apr;22(4):692-701. doi: 10.1038/mt.2013.285. Epub 2013 Dec 19. Mol Ther. 2014. PMID: 24352214 Free PMC article. Review. - Oligonucleotide-based strategies to combat polyglutamine diseases.
Fiszer A, Krzyzosiak WJ. Fiszer A, et al. Nucleic Acids Res. 2014 Jun;42(11):6787-810. doi: 10.1093/nar/gku385. Epub 2014 May 21. Nucleic Acids Res. 2014. PMID: 24848018 Free PMC article. Review. - RNA Interference of Human α-Synuclein in Mouse.
Kim YC, Miller A, Lins LC, Han SW, Keiser MS, Boudreau RL, Davidson BL, Narayanan NS. Kim YC, et al. Front Neurol. 2017 Jan 31;8:13. doi: 10.3389/fneur.2017.00013. eCollection 2017. Front Neurol. 2017. PMID: 28197125 Free PMC article. - Current trends in gene therapy to treat inherited disorders of the brain.
Matuszek Z, Brown BL, Yrigollen CM, Keiser MS, Davidson BL. Matuszek Z, et al. Mol Ther. 2025 May 7;33(5):1988-2014. doi: 10.1016/j.ymthe.2025.03.057. Epub 2025 Apr 2. Mol Ther. 2025. PMID: 40181540 Review. - Targeted gene silencing to induce permanent sterility.
Dissen GA, Lomniczi A, Boudreau RL, Chen YH, Davidson BL, Ojeda SR. Dissen GA, et al. Reprod Domest Anim. 2012 Aug;47 Suppl 4(Suppl 4):228-32. doi: 10.1111/j.1439-0531.2012.02080.x. Reprod Domest Anim. 2012. PMID: 22827375 Free PMC article.
References
- Krol J, Loedige I., and, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. - PubMed
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N., and, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC1 NS068099/NS/NINDS NIH HHS/United States
- T32 HL007121/HL/NHLBI NIH HHS/United States
- P01 NS050210/NS/NINDS NIH HHS/United States
- DK-54759/DK/NIDDK NIH HHS/United States
- NS-50210/NS/NINDS NIH HHS/United States
- NS-068099/NS/NINDS NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical