Comprehensive comparison of three commercial human whole-exome capture platforms - PubMed (original) (raw)
Comparative Study
doi: 10.1186/gb-2011-12-9-r95.
Yu Xu, Hui Jiang, Chris Tyler-Smith, Yali Xue, Tao Jiang, Jiawei Wang, Mingzhi Wu, Xiao Liu, Geng Tian, Jun Wang, Jian Wang, Huangming Yang, Xiuqing Zhang
Affiliations
- PMID: 21955857
- PMCID: PMC3308058
- DOI: 10.1186/gb-2011-12-9-r95
Comparative Study
Comprehensive comparison of three commercial human whole-exome capture platforms
Asan et al. Genome Biol. 2011.
Abstract
Background: Exome sequencing, which allows the global analysis of protein coding sequences in the human genome, has become an effective and affordable approach to detecting causative genetic mutations in diseases. Currently, there are several commercial human exome capture platforms; however, the relative performances of these have not been characterized sufficiently to know which is best for a particular study.
Results: We comprehensively compared three platforms: NimbleGen's Sequence Capture Array and SeqCap EZ, and Agilent's SureSelect. We assessed their performance in a variety of ways, including number of genes covered and capture efficacy. Differences that may impact on the choice of platform were that Agilent SureSelect covered approximately 1,100 more genes, while NimbleGen provided better flanking sequence capture. Although all three platforms achieved similar capture specificity of targeted regions, the NimbleGen platforms showed better uniformity of coverage and greater genotype sensitivity at 30- to 100-fold sequencing depth. All three platforms showed similar power in exome SNP calling, including medically relevant SNPs. Compared with genotyping and whole-genome sequencing data, the three platforms achieved a similar accuracy of genotype assignment and SNP detection. Importantly, all three platforms showed similar levels of reproducibility, GC bias and reference allele bias.
Conclusions: We demonstrate key differences between the three platforms, particularly advantages of solutions over array capture and the importance of a large gene target set.
Figures
Figure 1
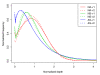
Normalized per-base sequencing-depth distribution on targets. For the purpose of comparison among the three platforms, we selected a set of reads with an average coverage of approximately 30-fold from each replicate. The depth and the frequency (the fraction of a certain depth-level bases for certain sequencing depth-coverage in the total sequencing data) were normalized by the average coverage depth of each replicate on targets. NA-r1 and NA-r2, NS-r1 and NS-r2, and AS-r1 and AS-r2 represent each of two replicates for NimbleGen Sequence Capture Arrays, NimbleGen SeqCap EZ and Agilent SureSelect, respectively.
Figure 2
Genotype sensitivity. (a) Genotype sensitivity of six replicates at 30× sequencing depth. (b) Genotype sensitivity as a function of sequencing depth. For the analyses, subsets of reads from two combined replicate datasets for each platform were randomly extracted at different average depths. NA, NS and AS represent NimbleGen Sequence Capture Arrays, NimbleGen SeqCap EZ and Agilent SureSelect, respectively, while r1 and r2 are two replicate experiments for each platform.
Figure 3
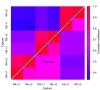
Correlation of sequencing depth and coverage rate on consensus targeted CCDSs. The graph shows pair-wise Pearson correlation coefficients for both sequencing depth (top-left triangle) and coverage rate (bottom-right triangle) based on the 182,259 CCDSs targeted by both Agilent and NimbleGen. NA, NS and AS represent NimbleGen Sequence Capture Arrays, NimbleGen SeqCap EZ and Agilent SureSelect, respectively, while r1 and r2 are two replicate experiments for each platform.
Similar articles
- Performance comparison of four commercial human whole-exome capture platforms.
Shigemizu D, Momozawa Y, Abe T, Morizono T, Boroevich KA, Takata S, Ashikawa K, Kubo M, Tsunoda T. Shigemizu D, et al. Sci Rep. 2015 Aug 3;5:12742. doi: 10.1038/srep12742. Sci Rep. 2015. PMID: 26235669 Free PMC article. - Comparison of solution-based exome capture methods for next generation sequencing.
Sulonen AM, Ellonen P, Almusa H, Lepistö M, Eldfors S, Hannula S, Miettinen T, Tyynismaa H, Salo P, Heckman C, Joensuu H, Raivio T, Suomalainen A, Saarela J. Sulonen AM, et al. Genome Biol. 2011 Sep 28;12(9):R94. doi: 10.1186/gb-2011-12-9-r94. Genome Biol. 2011. PMID: 21955854 Free PMC article. - Performance comparison of four exome capture systems for deep sequencing.
Chilamakuri CS, Lorenz S, Madoui MA, Vodák D, Sun J, Hovig E, Myklebost O, Meza-Zepeda LA. Chilamakuri CS, et al. BMC Genomics. 2014 Jun 9;15(1):449. doi: 10.1186/1471-2164-15-449. BMC Genomics. 2014. PMID: 24912484 Free PMC article. - A comparative analysis of exome capture.
Parla JS, Iossifov I, Grabill I, Spector MS, Kramer M, McCombie WR. Parla JS, et al. Genome Biol. 2011 Sep 29;12(9):R97. doi: 10.1186/gb-2011-12-9-r97. Genome Biol. 2011. PMID: 21958622 Free PMC article. - Types of array findings detectable in cytogenetic diagnosis: a proposal for a generic classification.
Srebniak MI, Diderich KE, Govaerts LC, Joosten M, Riedijk S, Galjaard RJ, Van Opstal D. Srebniak MI, et al. Eur J Hum Genet. 2014 Jul;22(7):856-8. doi: 10.1038/ejhg.2013.254. Epub 2013 Nov 6. Eur J Hum Genet. 2014. PMID: 24193341 Free PMC article. Review. No abstract available.
Cited by
- Novel Informatic Tools to Support Functional Annotation of the Durum Wheat Genome.
Fruzangohar M, Kalashyan E, Kalambettu P, Ens J, Wiebe K, Pozniak CJ, Tricker PJ, Baumann U. Fruzangohar M, et al. Front Plant Sci. 2019 Oct 10;10:1244. doi: 10.3389/fpls.2019.01244. eCollection 2019. Front Plant Sci. 2019. PMID: 31649706 Free PMC article. - Novel SLCO2A1compound heterozygous mutation causing primary hypertrophic osteoarthropathy with Bartter-like hypokalemia in a Chinese family.
Jiang Y, Du J, Song YW, Wang WB, Pang QQ, Li M, Wang O, Lian XL, Xing XP, Xia WB. Jiang Y, et al. J Endocrinol Invest. 2019 Oct;42(10):1245-1252. doi: 10.1007/s40618-019-01048-z. Epub 2019 Apr 19. J Endocrinol Invest. 2019. PMID: 31004291 - Spectrum of Genetic Variants Associated with Anterior Segment Dysgenesis in South Florida.
Thanikachalam S, Hodapp E, Chang TC, Swols DM, Cengiz FB, Guo S, Zafeer MF, Seyhan S, Bademci G, Scott WK, Grajewski A, Tekin M. Thanikachalam S, et al. Genes (Basel). 2020 Mar 26;11(4):350. doi: 10.3390/genes11040350. Genes (Basel). 2020. PMID: 32224865 Free PMC article. - Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy.
Golbus JR, Puckelwartz MJ, Dellefave-Castillo L, Fahrenbach JP, Nelakuditi V, Pesce LL, Pytel P, McNally EM. Golbus JR, et al. Circ Cardiovasc Genet. 2014 Dec;7(6):751-759. doi: 10.1161/CIRCGENETICS.113.000578. Epub 2014 Sep 1. Circ Cardiovasc Genet. 2014. PMID: 25179549 Free PMC article. - Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists.
Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, Temple-Smolkin RL, Voelkerding KV, Nikiforova MN. Jennings LJ, et al. J Mol Diagn. 2017 May;19(3):341-365. doi: 10.1016/j.jmoldx.2017.01.011. Epub 2017 Mar 21. J Mol Diagn. 2017. PMID: 28341590 Free PMC article. Review.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C. et al.Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. - DOI - PubMed
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N. et al.The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. - DOI - PubMed
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous