Direct lineage conversion of terminally differentiated hepatocytes to functional neurons - PubMed (original) (raw)
Direct lineage conversion of terminally differentiated hepatocytes to functional neurons
Samuele Marro et al. Cell Stem Cell. 2011.
Abstract
Several recent studies have showed that mouse and human fibroblasts can be directly reprogrammed into induced neuronal (iN) cells, bypassing a pluripotent intermediate state. However, fibroblasts represent heterogeneous mesenchymal progenitor cells that potentially contain neural crest lineages, and the cell of origin remained undefined. This raises the fundamental question of whether lineage reprogramming is possible between cell types derived from different germ layers. Here, we demonstrate that terminally differentiated hepatocytes can be directly converted into functional iN cells. Importantly, single-cell and genome-wide expression analyses showed that fibroblast- and hepatocyte-derived iN cells not only induced a neuronal transcriptional program, but also silenced their donor transcriptome. The remaining donor signature decreased over time and could not support functional hepatocyte properties. Thus, the reprogramming factors lead to a binary lineage switch decision rather than an induction of hybrid phenotypes, but iN cells retain a small but detectable epigenetic memory of their donor cells.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
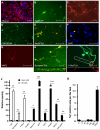
Figure 1. Induction of neuronal cells from liver cells
(A) Liver cells expressed albumin after isolation (red). (B) TauEGFP-positive cells with neuronal morphology (green, live image) and (C) Tuj1-positive (red) cells 13 and 22 days after dox induction, respectively. (D) PSA-NCAM (green) (E, F) NeuN (red) Tuj1 (green) (G) MAP2 and (H) synapsin-positive cells from liver cultures displayed complex neuronal morphologies. (I) A fraction of the Tuj1-positive cells expressed vGLUT1. (J) qPCR analysis of TauEGFP-positive FACS-sorted cells 21 days after dox (white) compared to liver cells 4 days after purification (black) (n=3). For neuronal markers the liver cell values were set to 1; for liver cell markers the TauEGFP-sorted cell values were set to 1. (K) Efficiency of generating liver-iN cells was quantified in 15 randomly picked 20x microscopic fields (n=3). Scale bars: 100μm (C), 50μm (A, B, D, E, H-I).
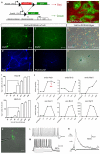
Figure 2. iN cells can be derived from terminally differentiated hepatocytes
(A) Experimental rationale of the genetic lineage tracing. (B) Albumin-Cre/Rosa26-mTmG liver cultures exhibited EGFP-positive hepatocytes and tdTomato-positive cells 5 days after isolation. (C-F) EGFP-positive Hep-iN cells derived 13 days after dox expressed Tuj1 and PSA-NCAM. (G) Hepatocyte cultures from Albumin-Cre/Rosa26-Bgeo mice showed β-galactosidase activity. (H) After infection both unstained and Xgal-stained neuronal like cells could be identified. (I) Quantification of Hep-iN and MEF-iN cells at day 23 after infection. Dox was removed from culture dishes at time points shown (n=3) (B) qPCR analysis for endogenous mRNA (endo) or viral mRNA (exo). Dox was withdrawn at day 12 (n=2) (K) EGFP-positive cell from Albumin-Cre/Rosa26-mTmG transgenic were characterized by patch clamping. (L) Spontaneous action potentials recorded in Hep-iN cells 30 days after dox induction (n=8). (M) Repetitive action potentials could be induced when increasing amounts of current was injected (n=16). (N) Evoked postsynaptic response recorded from a Hep-iN cell co-cultured with mouse cortical neurons. Arrow indicates time point of stimulation. Scale bars: 20μm (K), 25 μm (H) 50 μm (B, E, F, G), 100 μm (C, D).
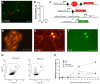
Figure 3. Efficiencies and timing of Hep-iN cell generation
(A) Most Tuj1-positive cells (green) were BrdU negative (red). (B) Quantification of BrdU/Tuj1 double-positive liver-iN cells after BrdU treatment from day 0–13 or day 1–13 after dox induction (n=3). (C) Lineage tracing outcome in Albumin-Cre/Rosa26-tdTomato/TauEGFP mice. (D) Freshly isolated hepatocytes (E,F) Hep-iN cells are double positive. (G) Representative FACS plots showing iN cells (EGFP) from hepatocytes (tdTomato positive) and non-hepatocyte (tdTomato negative) 13 days after dox. (H) Efficiency of conversion to iN cells in mouse embryonic fibroblasts (MEF ■), postnatal tail tip fibroblasts (TTF ▲), hepatocytes (□), non-hepatocyte liver cells (●) expressed as percentage of infected cells that successfully activate the TauEGFP reporter. Scale bars: 50μm (A, D, E, F).
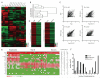
Figure 4. Global transcriptional remodeling during lineage conversion
(A) Heatmap of microarray data illustrating all differentially expressed genes among hepatocytes, d13 and d22 Hep-iN cells, MEF-iN and TTF-iN cells, d7 and d13 NPC-derived neurons and cortical neurons (12,275 genes). Expression levels are shown as mean centered log2 values. Red indicates up-regulated genes whereas green indicates down-regulated genes. The scale extends from 1.988- to 15.691-fold over mean (−2 to +2 in log2 space) as is indicated on the bottom. (B) Hierarchical clustering among all the samples analyzed. (C) Pearson’s distribution and correlation analysis of MEF vs. Hepatocytes, MEF-iN vs. Hep-iN at different stages of conversion and Hep-iN d22 vs. NPC-N d13. Shown are 1522 probes that are at least 4-fold differentially expressed. (D) Heatmaps showing the MEF and liver signature genes in donor and respective day 22 iN cells. (E) Single-cell gene expression profiling. Rows represent the evaluated genes and columns represent individual cells. Heat map represents the threshold Ct values. Table indicates percentage of positive cells in the three populations (H= hepatocytes, iN= Hep-iN cells, N= cortical neurons). (F) Distribution of hepatocytes, neurons, and Hep-iN cells as function of how many hepatic genes are expressed per single cell.
Similar articles
- Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq.
Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M, Quake SR. Treutlein B, et al. Nature. 2016 Jun 16;534(7607):391-5. doi: 10.1038/nature18323. Epub 2016 Jun 8. Nature. 2016. PMID: 27281220 Free PMC article. - JMJD3 aids in reprogramming of bone marrow progenitor cells to hepatic phenotype through epigenetic activation of hepatic transcription factors.
Kochat V, Equbal Z, Baligar P, Kumar V, Srivastava M, Mukhopadhyay A. Kochat V, et al. PLoS One. 2017 Mar 22;12(3):e0173977. doi: 10.1371/journal.pone.0173977. eCollection 2017. PLoS One. 2017. PMID: 28328977 Free PMC article. - Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors.
Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Huang P, et al. Nature. 2011 May 11;475(7356):386-9. doi: 10.1038/nature10116. Nature. 2011. PMID: 21562492 - Engineering cell fate: Spotlight on cell-activation and signaling-directed lineage conversion.
Ebrahimi B. Ebrahimi B. Tissue Cell. 2016 Oct;48(5):475-87. doi: 10.1016/j.tice.2016.07.005. Epub 2016 Jul 26. Tissue Cell. 2016. PMID: 27514850 Review. - Development-inspired reprogramming of the mammalian central nervous system.
Amamoto R, Arlotta P. Amamoto R, et al. Science. 2014 Jan 31;343(6170):1239882. doi: 10.1126/science.1239882. Science. 2014. PMID: 24482482 Free PMC article. Review.
Cited by
- New Insights Into the Intricacies of Proneural Gene Regulation in the Embryonic and Adult Cerebral Cortex.
Oproescu AM, Han S, Schuurmans C. Oproescu AM, et al. Front Mol Neurosci. 2021 Feb 15;14:642016. doi: 10.3389/fnmol.2021.642016. eCollection 2021. Front Mol Neurosci. 2021. PMID: 33658912 Free PMC article. Review. - Chemical transdifferentiation: closer to regenerative medicine.
Xu A, Cheng L. Xu A, et al. Front Med. 2016 Jun;10(2):152-65. doi: 10.1007/s11684-016-0445-z. Epub 2016 May 3. Front Med. 2016. PMID: 27142989 Review. - Nonviral direct conversion of primary mouse embryonic fibroblasts to neuronal cells.
Adler AF, Grigsby CL, Kulangara K, Wang H, Yasuda R, Leong KW. Adler AF, et al. Mol Ther Nucleic Acids. 2012 Jul 10;1(7):e32. doi: 10.1038/mtna.2012.25. Mol Ther Nucleic Acids. 2012. PMID: 23344148 Free PMC article. - Applications of Induced Pluripotent Stem Cells in Studying the Neurodegenerative Diseases.
Wan W, Cao L, Kalionis B, Xia S, Tai X. Wan W, et al. Stem Cells Int. 2015;2015:382530. doi: 10.1155/2015/382530. Epub 2015 Jul 9. Stem Cells Int. 2015. PMID: 26240571 Free PMC article. Review. - Direct conversion of dermal fibroblasts into neural progenitor cells by a novel cocktail of defined factors.
Tian C, Ambroz RJ, Sun L, Wang Y, Ma K, Chen Q, Zhu B, Zheng JC. Tian C, et al. Curr Mol Med. 2012 Feb;12(2):126-37. doi: 10.2174/156652412798889018. Curr Mol Med. 2012. PMID: 22172100 Free PMC article.
References
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet Beta cells. Cell Stem Cell. 2011;9:17–23. - PubMed
- Braeuning A, Ittrich C, Kohle C, Hailfinger S, Bonin M, Buchmann A, Schwarz M. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006;273:5051–5061. - PubMed
- Caiazzo M, Dell’anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011 - PubMed
- Horb ME, Shen CN, Tosh D, Slack JM. Experimental conversion of liver to pancreas. Curr Biol. 2003;13:105–115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- R01MH092931-01/MH/NIMH NIH HHS/United States
- R01 MH092931/MH/NIMH NIH HHS/United States
- RC4 NS073015/NS/NINDS NIH HHS/United States
- RC4 NS073015-01/NS/NINDS NIH HHS/United States
- R01 MH092931-01/MH/NIMH NIH HHS/United States
- RC4NS073015-01/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases