Global identification of modular cullin-RING ligase substrates - PubMed (original) (raw)
. 2011 Oct 14;147(2):459-74.
doi: 10.1016/j.cell.2011.09.019. Epub 2011 Sep 29.
Andrew E H Elia, Qikai Xu, Claudio R Thoma, Lior Izhar, Yumei Leng, Ailan Guo, Yi-Ning Chen, John Rush, Paul Wei-Che Hsu, Hsueh-Chi Sherry Yen, Stephen J Elledge
Affiliations
- PMID: 21963094
- PMCID: PMC3226719
- DOI: 10.1016/j.cell.2011.09.019
Global identification of modular cullin-RING ligase substrates
Michael J Emanuele et al. Cell. 2011.
Erratum in
- Cell. 2012 Feb 3;148(3):620-2
Abstract
Cullin-RING ligases (CRLs) represent the largest E3 ubiquitin ligase family in eukaryotes, and the identification of their substrates is critical to understanding regulation of the proteome. Using genetic and pharmacologic Cullin inactivation coupled with genetic (GPS) and proteomic (QUAINT) assays, we have identified hundreds of proteins whose stabilities or ubiquitylation status are regulated by CRLs. Together, these approaches yielded many known CRL substrates as well as a multitude of previously unknown putative substrates. We demonstrate that one substrate, NUSAP1, is an SCF(Cyclin F) substrate during S and G2 phases of the cell cycle and is also degraded in response to DNA damage. This collection of regulated substrates is highly enriched for nodes in protein interaction networks, representing critical connections between regulatory pathways. This demonstrates the broad role of CRL ubiquitylation in all aspects of cellular biology and provides a set of proteins likely to be key indicators of cellular physiology.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Figure 1. GPS Profiling and Chemical Genetic Inhibition of the Nedd8 Pathway Identifies CRL Substrates
(A) Schematic representation of a GPS screen. The GPS viral vector expresses a single transcript containing both DsRed and an EGFP-ORF, separated by an IRES. (i) A GPS cell library expressing each protein encoded by the Human ORFeome Collection was constructed, expressing a single GFP-ORF per cell. (ii) The library, with and without Cullin inhibition, is FACS sorted into bins based on its EGFP/DsRed ratio. (iii) Genomic DNA is isolated from sorted cells and ORFs are PCR amplified using primers that target the viral backbone. (iv) PCR amplified ORFs are transcribed, labeled and hybridized to DNA microarrays containing probes to each ORF. (v) Probes are analyzed and graphed across the bins to identify probe distributions that shift in response to CRL inhibition. (B) HeLa and 293T cells were treated for 4 h with increasing concentrations of MLN4924 (0, 0.1, 1 and 10 µM) and then immunoblotted for NRF2, CDT1 and Vinculin (loading control). (C) The indicated 293T GPS cell lines were treated with MLN4924 for 4 h and analyzed by flow cytometry. Histograms show the EGFP/DsRed ratio for cells expressing each of the specified ORFs or an empty vector control. (D) Left-Scatter plot of the PSI for each probe in the screen under the two conditions (DMSO vs. MLN4924). Right- Histogram of the ΔPSI for each probe analyzed in the screen. (E) Schematic representation of a subset of the known substrates that were identified in this screen. (F) 293T cells expressing the indicated EGFP tagged candidate substrate were immunoblotted to examine protein stabilization following 4 h treatment with MLN4924. (See also, Figure S1, S2, S3, S6 and Tables S1,S2 and S7)
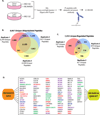
Figure 2. Mass Spectrometry Identification of CRL Substrates using QUAINT
(A) Schematic representation of the QUAINT experimental design to identify CRL regulated proteins. (B) Overlap between peptides identified in three QUAINT replicates. (C) Overlap between MLN4924 regulated peptides in the three QUAINT replicates. (D) Graphic depiction of the overlap between the MLN4924 GPS and QUAINT screens. Green represents known substrate adaptors; blue represents known substrates; red represents proteins with known interactions with CRL ligase components; purple are proteins that were identified in additional DN-Cul GPS screens; orange are proteins validated in this study by endogenous immunoblot. (See also Figure S2, S3, S6 and Tables S3 and S7)
Figure 3. GPS Screen to Identify CRL4 Substrates
(A) 293T cells expressing either FLAG-DN-Cul4 or an empty vector control were treated with UV or 4NQO to induce DNA damage. Vinculin was blotted a loading control. DN-Cul4 was detected using Flag antibodies. (B) 293T cells expressing GPS-CDKN1A were treated with UV light in the presence and absence of DN-Cul4. (C) A subset of validated candidate substrates that were tested in 293T cells using flow cytometry. (D) Functional category and protein family enrichment analysis for the proteins validating in the CRL4 GPS screen. (E) The overlap between CRL4 and MLN4924 GPS screens. (See also Figure S4 and S6 and Tables S4 and S7)
Figure 4. GPS Screen to Identify CRL3 Substrates
(A) 293T cells treated with empty vector or DN-Cul3 were immunoblotted for NRF2 and Ran (loading control). (B) 293T cells expressing GPS-CDKN1A (negative control), GPS-NRF2 or the GPS-library were treated with DN-Cul3 for 18 hours and analyzed by flow cytometry. (C) GPS-NRF2 cells were treated with DMSO or the oxidizing agent TBHQ for 4 h. (D) 293T cells expressing GPS-NRF2 that were treated with DN-Cul3 for increasing amounts of time were analyzed by flow cytometry. (E) Shown is a subset of validated candidate substrates that were tested in 293T cells using flow cytometry. (F) The overlap between CRL3 and MLN4924 GPS screens. (G) Functional category and protein family enrichment analysis for proteins overlap between the CRL3 and MLN4924 GPS screens. (See also Tables S5 and S7)
Figure 5. GPS Screen to Identify SCF Substrates
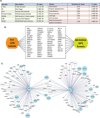
(A) Functional category and protein family enrichment analysis for the proteins that validated in the CRL4 GPS screen. (B) The overlap between CRL4 and MLN4924 GPS screens. (C) A sub-network demonstrating the high degree of betweenness for proteins regulated by the SCF. SCF candidate substrates are shown in cyan and circle size corresponds to the degree of connectivity within the network. (See also Figure S6 and Tables S6 and S7)
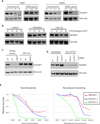
Figure 6. NUSAP1 is an SCFCyclin F Substrate
(A) 293T cells were treated with MLN4924 for 2, 4 and 8 h and immunblotted with antibodies to endogenous NUSAP1. (B) U2OS cells expressing the indicated DN-Cul were immunoblotted for NUSAP1. (C) U2OS cells were transfected with siRNA targeting firefly luciferase (siFF), or the F-box proteins Fbw7 and Cyclin F, incubated for 48h and immunoblotted for NUSAP1, Cyclin B and Vinculin (loading control). (D) 293T cells expressing GPS-NUSAP1 were treated with siFF or siCyclin F for 48 h and analyzed by flow cytometry. (E) HeLa cells were synchronized using a double thymidine block and released. Lysates were collected at the indicated times and immunoblotted for Phospho-Histone H3 on S10 (pH3S10- mitotic marker), Vinculin (loading control), PAF15, Cyclin B and NUSAP1. A semi-quantitative analysis of NUSAP1, Cyclin B and PAF15 protein levels (relative to loading controls), derived from the western blot shown, is graphed. (F) U2OS cells were synchronized using a double thymidine block and release. After the first thymidine block cells were transfected with siRNAs to Cyclin F or siFF. Following release cells were analyzed by immunoblot with the indiczated antibodies (* notes a cross reacting band in the Cyclin F immunoblot). A semi-quantitative analysis of NUSAP1 protein levels (relative to loading controls), derived from the western blot shown, is graphed. (G) Flag tagged Cyclin F or Cyclin F (1–270) was transfected into HeLa cells for 24 h. Cells were treated with 5 µM MG132 for 2 h, harvested and precipitated with anti-Flag agarose. The precipitates were immunoblotted for NUSAP1. (See also Figure S5)
Figure 7. NUSAP1 is Degraded Following UV and its Loss Sensitizes Cells to Anti-Tubulin Chemotherapeutics
(A) 293T and U2OS cells were treated with UV and collected at 1, 2 and 4 h for immunoblotting. Alternatively, the same cell lines were treated with either 0.2 or 10 µg/ml 4NQO and were harvested at 4 h. Cells were immunoblotted for NUSAP1 and Vinculin. (B) U2OS cells were treated with 0, 10, 25 or 50 J/m2 of UV and following treatment were incubated in media containing DMSO, 5 µM MLN4924 or 5 µM MG132. Four hours after UV treatment, cells were harvested for immunoblotting. (C) U2OS cells expressing Dn-Cul1 were treated with UV or 4QO and harvested after 4 h for immunoblot. (D) U2OS cells for transfected with siRNA targeting FF, Fbw7 or Cyclin F and after 48 h were treated with UV and harvested 4 h later for immunoblot. (E) U2OS cells were depleted of NUSAP1 using three independent siRNA for 24 hours, at which point cells were treated with taxol or nocodazole for an additional 72 h. Cell viability was assessed using cell titer glo. Survival of NUSAP1 depleted cells, relative to siFF treated controls, is reported at each concentrations tested. Each siRNA and drug combination was performed in triplicate and the graph reports the mean +/− the standard deviation. (See also Figure S5)
Similar articles
- Coupled monoubiquitylation of the co-E3 ligase DCNL1 by Ariadne-RBR E3 ubiquitin ligases promotes cullin-RING ligase complex remodeling.
Kelsall IR, Kristariyanto YA, Knebel A, Wood NT, Kulathu Y, Alpi AF. Kelsall IR, et al. J Biol Chem. 2019 Feb 22;294(8):2651-2664. doi: 10.1074/jbc.RA118.005861. Epub 2018 Dec 26. J Biol Chem. 2019. PMID: 30587576 Free PMC article. - Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells.
Luo Z, Pan Y, Jeong LS, Liu J, Jia L. Luo Z, et al. Autophagy. 2012 Nov;8(11):1677-9. doi: 10.4161/auto.21484. Epub 2012 Aug 9. Autophagy. 2012. PMID: 22874562 Free PMC article. - Introduction.
Sun Y. Sun Y. Adv Exp Med Biol. 2020;1217:1-8. doi: 10.1007/978-981-15-1025-0_1. Adv Exp Med Biol. 2020. PMID: 31898218 - Cullin-RING E3 Ubiquitin Ligases: Bridges to Destruction.
Nguyen HC, Wang W, Xiong Y. Nguyen HC, et al. Subcell Biochem. 2017;83:323-347. doi: 10.1007/978-3-319-46503-6_12. Subcell Biochem. 2017. PMID: 28271482 Free PMC article. Review. - Understanding cullin-RING E3 biology through proteomics-based substrate identification.
Harper JW, Tan MK. Harper JW, et al. Mol Cell Proteomics. 2012 Dec;11(12):1541-50. doi: 10.1074/mcp.R112.021154. Epub 2012 Sep 7. Mol Cell Proteomics. 2012. PMID: 22962057 Free PMC article. Review.
Cited by
- Quantifying ubiquitin signaling.
Ordureau A, Münch C, Harper JW. Ordureau A, et al. Mol Cell. 2015 May 21;58(4):660-76. doi: 10.1016/j.molcel.2015.02.020. Mol Cell. 2015. PMID: 26000850 Free PMC article. Review. - Identification of BBOX1 as a Therapeutic Target in Triple-Negative Breast Cancer.
Liao C, Zhang Y, Fan C, Herring LE, Liu J, Locasale JW, Takada M, Zhou J, Zurlo G, Hu L, Simon JM, Ptacek TS, Andrianov VG, Loza E, Peng Y, Yang H, Perou CM, Zhang Q. Liao C, et al. Cancer Discov. 2020 Nov;10(11):1706-1721. doi: 10.1158/2159-8290.CD-20-0288. Epub 2020 Jul 20. Cancer Discov. 2020. PMID: 32690540 Free PMC article. - A comprehensive method for detecting ubiquitinated substrates using TR-TUBE.
Yoshida Y, Saeki Y, Murakami A, Kawawaki J, Tsuchiya H, Yoshihara H, Shindo M, Tanaka K. Yoshida Y, et al. Proc Natl Acad Sci U S A. 2015 Apr 14;112(15):4630-5. doi: 10.1073/pnas.1422313112. Epub 2015 Mar 31. Proc Natl Acad Sci U S A. 2015. PMID: 25827227 Free PMC article. - Adaptive exchange sustains cullin-RING ubiquitin ligase networks and proper licensing of DNA replication.
Zhang Y, Jost M, Pak RA, Lu D, Li J, Lomenick B, Garbis SD, Li CM, Weissman JS, Lipford JR, Deshaies RJ. Zhang Y, et al. Proc Natl Acad Sci U S A. 2022 Sep 6;119(36):e2205608119. doi: 10.1073/pnas.2205608119. Epub 2022 Aug 29. Proc Natl Acad Sci U S A. 2022. PMID: 36037385 Free PMC article. - Ubiquitin-dependent regulation of COPII coat size and function.
Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgärtel C, Schekman R, Rape M. Jin L, et al. Nature. 2012 Feb 22;482(7386):495-500. doi: 10.1038/nature10822. Nature. 2012. PMID: 22358839 Free PMC article.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
- Benanti JA, Cheung SK, Brady MC, Toczyski DP. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat Cell Biol. 2007;9:1184–1191. - PubMed
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG011085/AG/NIA NIH HHS/United States
- R01 AG011085-19/AG/NIA NIH HHS/United States
- R01 GM044664/GM/NIGMS NIH HHS/United States
- R01 GM044664-13/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials