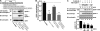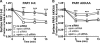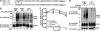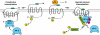Adaptor protein complex-2 (AP-2) and epsin-1 mediate protease-activated receptor-1 internalization via phosphorylation- and ubiquitination-dependent sorting signals - PubMed (original) (raw)
Adaptor protein complex-2 (AP-2) and epsin-1 mediate protease-activated receptor-1 internalization via phosphorylation- and ubiquitination-dependent sorting signals
Buxin Chen et al. J Biol Chem. 2011.
Abstract
Signaling by protease-activated receptor-1 (PAR1), a G protein-coupled receptor (GPCR) for thrombin, is regulated by desensitization and internalization. PAR1 desensitization is mediated by β-arrestins, like most classic GPCRs. In contrast, internalization of PAR1 occurs through a clathrin- and dynamin-dependent pathway independent of β-arrestins. PAR1 displays two modes of internalization. Constitutive internalization of unactivated PAR1 is mediated by the clathrin adaptor protein complex-2 (AP-2), where the μ2-adaptin subunit binds directly to a tyrosine-based motif localized within the receptor C-tail domain. However, AP-2 depletion only partially inhibits agonist-induced internalization of PAR1, suggesting a function for other clathrin adaptors in this process. Here, we now report that AP-2 and epsin-1 are both critical mediators of agonist-stimulated PAR1 internalization. We show that ubiquitination of PAR1 and the ubiquitin-interacting motifs of epsin-1 are required for epsin-1-dependent internalization of activated PAR1. In addition, activation of PAR1 promotes epsin-1 de-ubiquitination, which may increase its endocytic adaptor activity to facilitate receptor internalization. AP-2 also regulates activated PAR1 internalization via recognition of distal C-tail phosphorylation sites rather than the canonical tyrosine-based motif. Thus, AP-2 and epsin-1 are both required to promote efficient internalization of activated PAR1 and recognize discrete receptor sorting signals. This study defines a new pathway for internalization of mammalian GPCRs.
Figures
FIGURE 1.
Activated PAR1 colocalization with AP-2 and epsin-1. HeLa cells expressing PAR1 were left untreated (0 min) or treated with 100 μ
m
SFLLRN for 1 min at 37 °C. Cells were processed and imaged by confocal microscopy. Colocalization is revealed by the yellow color in the merged image. The images are representative of many cells examined in three different experiments. Insets represent magnifications of the boxed areas. Scale bar, 10 μm.
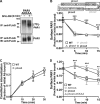
FIGURE 2.
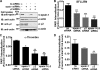
Activated PAR1 internalization is regulated by AP-2 and epsin-1. A, HeLa cells expressing PAR1 were transfected with 50 n
m
nonspecific (ns), epsin-1, μ2-adaptin, or epsin-1/μ2-adaptin siRNAs, lysed, and protein lysates were immunoblotted (IB) with the indicated antibodies. B and C, PAR1 expressing HeLa cells transfected with various siRNAs were incubated with 100 μ
m
SFLLRN or 10 n
m
α-thrombin for 10 min at 37 °C, fixed, and the amount of receptor remaining at the cell surface was measured by ELISA. The data (mean ± S.D., n = 3) are expressed as the fraction of cell surface antibody bound at 0 min and are representative of at least five independent experiments. The differences observed in PAR1 internalization were significant as determined by one-way ANOVA and Dunnett's multiple comparison tests (*, p < 0.05; **, p < 0.01; ***, p < 0.001). D, PAR1 expressing HeLa cells transfected with nonspecific, epsin-1, or μ2-adaptin siRNAs were prelabeled with M1 anti-PAR1 antibody for 1 h at 4 °C. Cells were washed, warmed to 37 °C, and incubated with serum-free media for 10 min. Antibody that remained bound to the cell surface was removed by washing three times with PBS (Ca2+ and Mg2+ free) containing 0.04% EDTA; M1 anti-FLAG antibody binding requires Ca2+. Cells were then lysed in 1% Triton X-100, 50 m
m
Tris-HCl, pH 7.4, 100 m
m
NaCl, 5 m
m
EDTA, and 3% BSA and the amount of internalized antibody-bound to receptor was quantified by ELISA. The data are expressed as a fraction of the initial antibody bound to the cell surface at 0 min at 4 °C before stripping. These data (mean ± S.D., n = 3) are representative of multiple independent experiments. The extent of PAR1 constitutive internalization in nonspecific versus μ2-adaptin siRNA-transfected cells was significant (**, p < 0.01), determined as described above.
FIGURE 3.
Agonist-induced internalization of endogenous PAR1 is mediated by epsin-1 and AP-2. A, human endothelial EA.hy926 cells were electroporated with 50 n
m
nonspecific (ns), epsin-1, μ2-adaptin, or epsin-1/μ2-adaptin siRNAs and lysates were immunoblotted (IB) with the indicated antibodies. B, endothelial cells were stimulated with 100 μ
m
TFLLRNPNDK for 30 min at 37 °C, and the amount of endogenous PAR1 internalization was determined by ELISA. The data (mean ± S.D., n = 3) are representative of three separate experiments. The differences in PAR1 internalization were significant as determined by one-way ANOVA and Dunnett's multiple comparison tests (*, p < 0.05; **, p < 0.01). C, endothelial cells were incubated with 100 μ
m
TFLLRNPNDK for 30 min at 37 °C, processed, and imaged by confocal microscopy. The images shown are representative of many cells examined in multiple independent experiments. Scale bar, 10 μm. Note that the nuclear staining is not specific to the epsin-1 antibody.
FIGURE 4.
Activated PAR1 internalization is mediated by epsin-1 UIMs and de-ubiquitination epsin-1. A, the schematic represents the various pSLIK vectors. Cells were incubated with 0.1–1 μg/ml of doxycycline (DOX) for 72 h at 37 °C. Cell lysates were immunoblotted (IB) with an anti-epsin-1 antibody (upper panel), anti-c-Myc antibody to detect Myc-tagged epsin-1 (middle panel), or anti-β-actin antibody to control for loading (lower panel). B, HeLa cells stably expressing pSLIK vectors encoding nonspecific (ns)- or epsin-1-shRNA alone, or co-expressing shRNA-resistant rat Myc-tagged epsin-1 or S195D,S220D,S245D UIM mutant were transfected with FLAG-tagged PAR1 wild-type. Cells were incubated with 100 μ
m
SFLLRN for 10 min at 37 °C, fixed, and the amount of cell surface receptor was measured by ELISA. The data (mean ± S.D., n = 3) are expressed as the fraction of the initial cell surface antibody bound at 0 min and is representative of four separate experiments. The differences in PAR1 internalization were significant as determined by one-way ANOVA and Dunnett's multiple comparison test (**, p < 0.01). C, HEK293 cells expressing HA-tagged PAR1 wild-type, FLAG-tagged ubiquitin, and Myc-tagged rat epsin-1 were stimulated with 100 μ
m
SFLLRN (+SF) or 2 μ
m
A23187 (A2), calcium ionophore, for 3 or 5 min at 37 °C. Cells were lysed, immunoprecipitated (IP), and epsin ubiquitination was detected. Similar findings were observed in three independent experiments and quantitated by densitometry. The data (mean ± S.D., n = 3) are expressed as the fraction of basal epsin-1 ubiquitination detected at 0 min. The differences in epsin-1 ubiquitination were significant as determined by one-way ANOVA and Dunnett's multiple comparison tests (**, p < 0.01).
FIGURE 5.
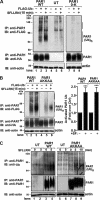
PAR1 displays basal and agonist-induced ubiquitination. A, HEK293 cells expressing HA-tagged PAR1 wild-type (WT), lysine-less 0-K mutant, and untransfected (UT) control were either mock transfected (−) or transfected with FLAG-tagged ubiquitin (+). Serum-starved cells were then incubated with or without 100 μ
m
SFLLRN for 15 min at 37 °C, immunoprecipitated (IP) with monoclonal anti-PAR1 antibody, and ubiquitination was detected with an anti-FLAG antibody. The membrane was then probed with an anti-HA antibody to detect PAR1 expression or an anti-actin antibody to control for equivalent amounts of cell lysates. These results are representative of at least three independent experiments. B, HEK293 cells expressing HA-PAR1 wild-type or AKKAA mutant were mock transfected (−) or transfected with FLAG-ubiquitin (+). Cells deprived of serum were then either left untreated or treated with 100 μ
m
SFLLRN for 15 min at 37 °C, immunoprecipitated, and PAR1 ubiquitination was detected using anti-FLAG antibodies. Membranes were stripped and reprobed with anti-HA antibody to detect PAR1 or anti-actin as a control. The amount of PAR1 ubiquitination detected (mean ± S.D., n = 3) is expressed as the fold over untreated control (Ctrl) from three different experiments. The differences in PAR1 WT and AKKAA ubiquitination were significant as determined by Student's t test (***, p < 0.001). C, HeLa cells expressing FLAG-tagged PAR1 WT, AKKAA mutant, and untransfected (UT) controls were stimulated with 100 μ
m
SFLLRN for various times at 37 °C and immunoprecipitated with anti-PAR1 antibody. Endogenous ubiquitination of PAR1 was detected by immunoblotting (IB) using an anti-ubiquitin antibody. Membranes were stripped and reprobed with an anti-PAR1 antibody to detect PAR1 or anti-actin antibody to control for total amounts of cell lysates. Similar findings were observed in three independent experiments.
FIGURE 6.
PAR1 ubiquitination is required for epsin-1-mediated internalization. HeLa cells expressing (A) PAR1 0-K or (B) PAR1 AKKAA mutant were transfected with nonspecific (ns) (□), epsin-1 (○), μ2-adaptin (◊), or epsin-1/μ2-adaptin (△) specific siRNAs. Serum-starved cells were then incubated with 100 μ
m
SFLLRN for varying times at 37 °C and the amount of receptor internalized was assayed by ELISA. The amount of PAR1 remaining on the cell surface (mean ± S.D., n = 3) are expressed as the fraction of receptor at the cell surface at 0 min and are representative of at least three independent experiments. The differences in PAR1 internalization observed in various siRNA-transfected cells was significant as determined by two-way ANOVA and Bonferroni post-tests (***, p < 0.001).
FIGURE 7.
PAR1 phosphorylation is critical for internalization and ubiquitination. A, PAR1 wild-type (WT) C-tail amino acid sequence and the distal tyrosine motif is underlined. Ubiquitination-deficient PAR1 0-K mutant has all intracytosolic lysines (K) converted to arginine (R) and is indicated as “K → R”. The phosphorylation-deficient mutant has all C-tail serines (S) and threonines (T) mutated to alanines (A), and indicated as Ser/Thr → Ala. B, HeLa cells expressing FLAG-tagged PAR1 WT, 0-K, or Ser/Thr → Ala mutants labeled with [32P]orthophosphate were stimulated with 100 μ
m
SFLLRN for 3 min at 37 °C and immunoprecipitated (IP) with anti-FLAG antibody. PAR1 labeled with 32P was visualized by autoradiography. The membrane was re-probed with anti-FLAG antibody to detect PAR1 expression. C, HeLa cells expressing the PAR1 Ser/Thr → Ala mutant were transfected with nonspecific (ns) (□), epsin-1 (○), μ2-adaptin (◊), or epsin-1/μ2-adaptin (△) specific siRNAs and then incubated with 100 μ
m
SFLLRN for varying times at 37 °C and the amount of receptor internalized was assayed by ELISA. The amount of PAR1 remaining on the cell surface (mean ± S.D., n = 3) are expressed as the fraction of receptor at the cell surface at 0 min and are representative of at least three independent experiments. D, HEK293 cells stably expressing the HA-PAR1 Ser/Thr → Ala mutant were transfected without (−) or with (+) FLAG-ubiquitin. Cells deprived of serum were then either left untreated or treated with 100 μ
m
SFLLRN for 15 min at 37 °C, immunoprecipitated, and PAR1 ubiquitination was detected using anti-FLAG antibodies. Membranes were reprobed with an anti-HA antibody to PAR1 or anti-actin to control for total amounts of cell lysates.
FIGURE 8.
Phosphorylation of the distal C-tail is important for AP-2-mediated activated PAR1 internalization. A, HeLa cells expressing FLAG-PAR1 AKKAA mutant labeled with [32P]orthophosphate were incubated with or without 100 μ
m
SFLLRN for 3 min at 37 °C and equivalent amounts of lysates were immunoprecipitated (IP) with anti-FLAG antibody. Immunoprecipitates were resolved by SDS-PAGE, transferred to PVDF membrane, and the amount of 32P-labeled PAR1 was visualized by autoradiography. The membrane was re-probed with anti-FLAG antibody to detect PAR1 expression. B, amino acid sequence of PAR1 phosphorylation-deficient mutants in which the proximal phos1, middle phos2, or distal phos3 C-tail serines and threonines were mutated to alanines. HeLa cells expressing FLAG-PAR1 wild-type (WT) (□), phos1 (△), phos2 (▿), or phos3 (◊) mutants were incubated with 100 μ
m
SFLLRN for varying times at 37 °C and the amount of receptor internalized was assayed by ELISA. The amount of PAR1 remaining on the cell surface (mean ± S.D., n = 3) is expressed as the fraction of receptor at the cell surface at 0 min and are representative of at least three independent experiments. The differences in PAR1 internalization observed with phos3 mutants versus WT was significant as determined by two-way ANOVA and Bonferroni post-tests (***, p < 0.001). C, HeLa cells expressing PAR1 wild-type (WT) or phos3 cluster mutant were prelabeled with M1 anti-PAR1 antibody for 1 h at 4 °C. Cells were washed, warmed to 37 °C, and incubated with serum-free media only for various times. Antibody that remained bound to the cell surface was removed by washing three times with PBS (Ca2+ and Mg2+ free) containing 0.04% EDTA; M1 anti-FLAG antibody binding requires Ca2+. Cells were then lysed in 1% Triton X-100, 50 m
m
Tris-HCl, pH 7.4, 100 m
m
NaCl, 5 m
m
EDTA, and 3% BSA and the amount of internalized antibody bound to receptor was quantified by ELISA. The data are expressed as a fraction of the initial antibody bound to the cell surface at 0 min at 4 °C before stripping. These data (mean ± S.D., n = 3) are representative of several independent experiments. D, HeLa cells expressing FLAG-PAR1 WT (□), AKKAA (○), phos3 (◊), or phos3-AKKAA (△) mutants were incubated with 100 μ
m
SFLLRN for varying times at 37 °C and the amount of receptor internalized was assayed by ELISA. The differences in PAR1 internalization observed with various mutants was significant as determined by two-way ANOVA and Bonferroni post-tests (**, p < 0.01; ***, p < 0.001).
FIGURE 9.
Model of PAR1 internalization. PAR1 displays two modes of internalization. Unactivated PAR1 internalizes constitutively through a mechanism mediated by the μ2-adaptin subunit of AP-2 binding to a tyrosine-based motif localized in the distal C terminus of PAR1. PAR1 is also basally ubiquitinated. Basal ubiquitination of unactivated PAR1 does function as an internalization signal and appears to negatively regulate constitutive internalization. In contrast, agonist stimulation of PAR1 results in rapid phosphorylation. Activated ubiquitinated and phosphorylated PAR1 is then recognized by epsin-1 and mediates internalization through receptor ubiquitin interaction with the UIMs of epsin-1. Moreover, epsin-1 is de-ubiquitinated after activation of PAR1, which may also increase its endocytic activity to facilitate internalization of activated ubiquitinated and phosphorylated PAR1. AP-2 is also critical for activated PAR1 internalization and appears to require phosphorylation at the receptor distal C terminus, rather than the tyrosine-based motif. The exact order and timing of epsin-1 and AP-2 recruitment to activated PAR1 is not known. It is also not known if epsin-1 and AP-2 interact with each other to coordinate activated PAR1 internalization through clathrin-coated pits.
Similar articles
- Palmitoylation of protease-activated receptor-1 regulates adaptor protein complex-2 and -3 interaction with tyrosine-based motifs and endocytic sorting.
Canto I, Trejo J. Canto I, et al. J Biol Chem. 2013 May 31;288(22):15900-12. doi: 10.1074/jbc.M113.469866. Epub 2013 Apr 11. J Biol Chem. 2013. PMID: 23580642 Free PMC article. - Regulation of protease-activated receptor 1 signaling by the adaptor protein complex 2 and R4 subfamily of regulator of G protein signaling proteins.
Chen B, Siderovski DP, Neubig RR, Lawson MA, Trejo J. Chen B, et al. J Biol Chem. 2014 Jan 17;289(3):1580-91. doi: 10.1074/jbc.M113.528273. Epub 2013 Dec 2. J Biol Chem. 2014. PMID: 24297163 Free PMC article. - Ubiquitination differentially regulates clathrin-dependent internalization of protease-activated receptor-1.
Wolfe BL, Marchese A, Trejo J. Wolfe BL, et al. J Cell Biol. 2007 Jun 4;177(5):905-16. doi: 10.1083/jcb.200610154. Epub 2007 May 29. J Cell Biol. 2007. PMID: 17535965 Free PMC article. - Signal transduction by protease-activated receptors.
Soh UJ, Dores MR, Chen B, Trejo J. Soh UJ, et al. Br J Pharmacol. 2010 May;160(2):191-203. doi: 10.1111/j.1476-5381.2010.00705.x. Br J Pharmacol. 2010. PMID: 20423334 Free PMC article. Review. - Signals for sorting of transmembrane proteins to endosomes and lysosomes.
Bonifacino JS, Traub LM. Bonifacino JS, et al. Annu Rev Biochem. 2003;72:395-447. doi: 10.1146/annurev.biochem.72.121801.161800. Epub 2003 Mar 6. Annu Rev Biochem. 2003. PMID: 12651740 Review.
Cited by
- Protease-activated Receptor-4 Signaling and Trafficking Is Regulated by the Clathrin Adaptor Protein Complex-2 Independent of β-Arrestins.
Smith TH, Coronel LJ, Li JG, Dores MR, Nieman MT, Trejo J. Smith TH, et al. J Biol Chem. 2016 Aug 26;291(35):18453-64. doi: 10.1074/jbc.M116.729285. Epub 2016 Jul 11. J Biol Chem. 2016. PMID: 27402844 Free PMC article. - Protease-activated receptors in health and disease.
Peach CJ, Edgington-Mitchell LE, Bunnett NW, Schmidt BL. Peach CJ, et al. Physiol Rev. 2023 Jan 1;103(1):717-785. doi: 10.1152/physrev.00044.2021. Epub 2022 Jul 28. Physiol Rev. 2023. PMID: 35901239 Free PMC article. Review. - Genetic Factors of the Disease Course After Sepsis: Rare Deleterious Variants Are Predictive.
Taudien S, Lausser L, Giamarellos-Bourboulis EJ, Sponholz C, Schöneweck F, Felder M, Schirra LR, Schmid F, Gogos C, Groth S, Petersen BS, Franke A, Lieb W, Huse K, Zipfel PF, Kurzai O, Moepps B, Gierschik P, Bauer M, Scherag A, Kestler HA, Platzer M. Taudien S, et al. EBioMedicine. 2016 Oct;12:227-238. doi: 10.1016/j.ebiom.2016.08.037. Epub 2016 Sep 15. EBioMedicine. 2016. PMID: 27639823 Free PMC article. - Palmitoylation of protease-activated receptor-1 regulates adaptor protein complex-2 and -3 interaction with tyrosine-based motifs and endocytic sorting.
Canto I, Trejo J. Canto I, et al. J Biol Chem. 2013 May 31;288(22):15900-12. doi: 10.1074/jbc.M113.469866. Epub 2013 Apr 11. J Biol Chem. 2013. PMID: 23580642 Free PMC article. - Site-specific polyubiquitination differentially regulates parathyroid hormone receptor-initiated MAPK signaling and cell proliferation.
Zhang Q, Xiao K, Liu H, Song L, McGarvey JC, Sneddon WB, Bisello A, Friedman PA. Zhang Q, et al. J Biol Chem. 2018 Apr 13;293(15):5556-5571. doi: 10.1074/jbc.RA118.001737. Epub 2018 Feb 14. J Biol Chem. 2018. PMID: 29444827 Free PMC article.
References
- Oldham W. M., Hamm H. E. (2008) Nat. Rev. Mol. Cell Biol. 9, 60–71 - PubMed
- Edeling M. A., Smith C., Owen D. (2006) Nat. Rev. Mol. Cell Biol. 7, 32–44 - PubMed
- Traub L. M. (2009) Nat. Rev. Mol. Cell Biol. 10, 583–596 - PubMed
- Moore C. A., Milano S. K., Benovic J. L. (2007) Annu. Rev. Physiol. 69, 451–482 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials