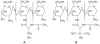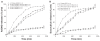Thiolated chitosan nanoparticles as a delivery system for antisense therapy: evaluation against EGFR in T47D breast cancer cells - PubMed (original) (raw)
Thiolated chitosan nanoparticles as a delivery system for antisense therapy: evaluation against EGFR in T47D breast cancer cells
Fatemeh Talaei et al. Int J Nanomedicine. 2011.
Retraction in
- Thiolated Chitosan Nanoparticles as a Delivery System for Antisense Therapy: Evaluation against EGFR in T47D Breast Cancer Cells [Retraction].
[No authors listed] [No authors listed] Int J Nanomedicine. 2022 Aug 11;17:3581-3582. doi: 10.2147/IJN.S385585. eCollection 2022. Int J Nanomedicine. 2022. PMID: 35983481 Free PMC article.
Abstract
Thiolated chitosan has high transfection and mucoadhesive properties. We investigated the potential of two recently synthesized polymers: NAC-C (N-acetyl cysteine-chitosan) and NAP-C (N-acetyl penicillamine-chitosan) in anticancer drug delivery targeting epidermal growth factor receptor (EGFR). Doxorubicin (DOX) and antisense oligonucleotide (ASOND)-loaded polymer nanoparticles were prepared in water by a gelation process. Particle characterization, drug loading, and drug release were evaluated. To verify drug delivery efficiency in vitro experiments on a breast cancer cell line (T47D) were performed. EGFR gene and protein expression was analyzed by real time quantitative polymerase chain reaction and Western blotting, respectively. A loading percentage of 63% ± 5% for ASOND and 70% ± 5% for DOX was achieved. Drug release data after 15 hours showed that ASOND and DOX were completely released from chitosan-based particles while a lower and more sustained release of only 22% ± 8% was measured for thiolated particles. In a cytosol simulated release medium/reducing environment, such as found intracellularly, polymer-based nanoparticles dissociated, liberating approximately 50% of both active substances within 7 hours. ASOND-loaded polymer nanoparticles had higher stability and high mucoadhesive properties. The ASOND-loaded thiolated particles significantly suppressed EGFR gene expression in T47D cells compared with ASOND-loaded chitosan particles and downregulated EGFR protein expression in cells. This study could facilitate future investigations into the functionality of NAP-C and NAC-C polymers as an efficient ASOND delivery system in vitro and in vivo.
Keywords: T47D breast cancer cells; antisense oligonucleotide; controlled release; doxorubicin; epidermal growth factor receptor; nanoparticles; thiolated chitosan.
Figures
Figure 1
Presumptive structures of (A) NAC-chitosan and (B) NAP-chitosan conjugates. Abbreviations: NAC**,** N-acetyl cysteine; NAP, N-acetyl penicillamine.
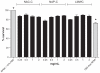
Figure 2
The effect of different concentration (0.25, 0.5, 1, 2 mg/mL) of polymers (NAC-C, NAP-C, LMWC) on T47D cancer cell proliferation 48 hours after treatment compared with positive control (RPMI + 10% FBS) and negative control (FBS-free RPMI). The data are mean ± SD of three experiments in four wells. In ANOVA tests all the data were compared to controls demonstrating a significant difference (P < 0.0001). Note: *Significantly different from all other groups. Abbreviations: ANOVA, analysis of variance; NAC-C, N-acetyl cysteine-chitosan; NAP-C, N-acetyl penicillamine-chitosan; LMWC, low-molecular-weight chitosan; RPMI, Roswell Park Memorial Institute medium; FBS, fetal bovine serum; SD, standard deviation.
Figure 3
Representative SEM micrograph of nanoparticles formulated via sulfate gelation with Na2SO4. (A) chitosan nanoparticles, (B) thiolated chitosan (NAP-C) nanoparticles, (C) DOX-NAC-C nanoparticles, (D) ASOND-NAC-C nanoparticles demonstrate a spherical, uniform shape with a particle size of 150–300 nm. Abbreviations: DOX, doxorubicin; NAC-C, N-acetyl cysteine-chitosan; NAP-C, N-acetyl penicillamine-chitosan; ASOND, antisense oligonucleotide; SEM, scanning electron microscope.
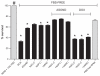
Figure 4
MTS assay measured the effect of polymer nanoparticles (0.5 mg/mL) containing ASOND and DOX (25 × 10−7 M) on T47D cancer cell proliferation 24 hours after treatment. The data are mean ± SD of three separate experiments in four wells. In ANOVA tests all the data were compared with controls (RPMI + 10% FBS- and FBS-free RPMI). **Notes: ***Significantly different from FBS-free RPMI; #RPMI + 10% FBS is significantly different from all the other treatments (P < 0.0001). Abbreviations: ASOND, antisense oligonucleotide; DOX, doxorubicin; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, NAC-C, N-acetyl cysteine-chitosan; NAP-C, N-acetyl penicillamine-chitosan; LMWC, low-molecular-weight chitosan; RPMI, Roswell Park Memorial Institute medium; FBS, fetal bovine serum; SD, standard deviation.
Figure 5
Percentage release of ASOND and doxorubicin from 5 mg of drug-loaded nanoparticles during 15 hours in PBS and cytosol simulated medium using DTT. (A) Percentage release of ASOND, (B) Percentage release of doxorubicin. The release data are mean ± SD for each formulation at different time points indicated (n = 4). Abbreviations: ASOND**,** antisense oligonucleotide; DOX, doxorubicin; NAC-C, N-acetyl cysteine-chitosan; NAP-C, N-acetyl penicillamine-chitosan; LMWC, low-molecular- weight chitosan; RPMI, Roswell Park Memorial Institute medium; FBS, fetal bovine serum; PBS, phosphate buffered saline; DTT, threo-1,4-dimercapto-2,3- butandiol; SD, standard deviation.
Figure 6
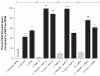
Real-time PCR analysis of EG FR gene expression in T47D cells subjected to different treatments. The results were normalized to beta-actin gene expression and the relative expression of EGFR was presented as percent knock down of EGFR. FBS-free RPMI treatment was chosen as control, which presented no knock down of EGFR and all the other cell treatments were compared with this control. All data are presented as percent knock down of EGFR and are significantly different to control. **Notes: ***Difference to control or free polymers (gray bars) within each group separated by indicator lines; #significant difference of each formulation containing ASOND or DOX compared to free DOX or ASOND (P < 0.05); data are mean SD (n = 3). Abbreviations: ASOND, antisense oligonucleotide; DOX, doxorubicin; EGFR, epidermal growth factor receptor; NAC-C, N-acetyl cysteine-chitosan; NAP-C, N-acetyl penicillamine-chitosan; LMWC, low-molecular-weight chitosan; RPMI, Roswell Park Memorial Institute medium; FBS, fetal bovine serum; PBS, phosphate buffered saline; SD, standard deviation.
Figure 7
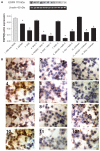
Analysis of EGFR protein expression in T47D cells. (A) Western blot analysis of EGFR expression in T47D cells through different treatments; (B) immunocytochemical analysis of EGFR protein expression in T47D cells. (1) FBS-free RPMI; (2) ASOND; (3) DOX; (4) NAC-C; (5) ASOND + NAC-C; (6) DOX + NAC-C;, (7) NAP-C; (8) ASOND + NAP-C; (9) DOX + NAP-C; (10) Chitosan; (11) ASOND + chitosan; (12) DOX + chitosan. The brown stain signifies higher expression of this protein which is observed in cytoplasm and on the membrane of the cells and the blue stain indicates the nuclei of the cell. Note: *Significantly different from FBS-free RPMI treated cells as controls (mean ± SD; n = 3). Abbreviations: ASOND, antisense oligonucleotide; DOX, doxorubicin; EGFR, epidermal growth factor receptor; NAC-C, N-acetyl cysteine-chitosan; NAP-C, N-acetyl penicillamine-chitosan; LMWC, low-molecular-weight chitosan; RPMI, Roswell Park Memorial Institute medium; FBS, fetal bovine serum; PBS, phosphate buffered saline; SD, standard deviation.
Similar articles
- Preparation of Modified Chitosan-based Nanoparticles for Efficient Delivery of Doxorubicin and/or Cisplatin to Breast Cancer Cells.
Matalqah SM, Aiedeh K, Mhaidat NM, Alzoubi KH, Al-Husein BA. Matalqah SM, et al. Curr Cancer Drug Targets. 2022;22(2):133-141. doi: 10.2174/1568009622666220126100532. Curr Cancer Drug Targets. 2022. PMID: 35081892 - Development of anti-HER2-targeted doxorubicin-core-shell chitosan nanoparticles for the treatment of human breast cancer.
Naruphontjirakul P, Viravaidya-Pasuwat K. Naruphontjirakul P, et al. Int J Nanomedicine. 2019 Jun 4;14:4105-4121. doi: 10.2147/IJN.S198552. eCollection 2019. Int J Nanomedicine. 2019. PMID: 31239670 Free PMC article. - Chitosan-thioglycolic acid conjugate: an alternative carrier for oral nonviral gene delivery?
Martien R, Loretz B, Thaler M, Majzoob S, Bernkop-Schnürch A. Martien R, et al. J Biomed Mater Res A. 2007 Jul;82(1):1-9. doi: 10.1002/jbm.a.31135. J Biomed Mater Res A. 2007. PMID: 17265441 - Plausible role of chitosan in drug and gene delivery against resistant breast cancer cells.
Nandgude T, Pagar R. Nandgude T, et al. Carbohydr Res. 2021 Aug;506:108357. doi: 10.1016/j.carres.2021.108357. Epub 2021 Jun 4. Carbohydr Res. 2021. PMID: 34146935 Review. - Stimuli-sensitive Chitosan-based Nanosystems-immobilized Nucleic Acids for Gene Therapy in Breast Cancer and Hepatocellular Carcinoma.
Naghib SM, Ahmadi B, Mozafari MR. Naghib SM, et al. Curr Top Med Chem. 2024;24(17):1464-1489. doi: 10.2174/0115680266293173240506054439. Curr Top Med Chem. 2024. PMID: 38752630 Review.
Cited by
- Critical evaluation of biodegradable polymers used in nanodrugs.
Marin E, Briceño MI, Caballero-George C. Marin E, et al. Int J Nanomedicine. 2013;8:3071-90. doi: 10.2147/IJN.S47186. Epub 2013 Aug 19. Int J Nanomedicine. 2013. PMID: 23990720 Free PMC article. Review. - Downregulation of CD73 in 4T1 breast cancer cells through siRNA-loaded chitosan-lactate nanoparticles.
Jadidi-Niaragh F, Atyabi F, Rastegari A, Mollarazi E, Kiani M, Razavi A, Yousefi M, Kheshtchin N, Hassannia H, Hadjati J, Shokri F. Jadidi-Niaragh F, et al. Tumour Biol. 2016 Jun;37(6):8403-12. doi: 10.1007/s13277-015-4732-0. Epub 2016 Jan 5. Tumour Biol. 2016. PMID: 26733167 - Thiolated Nanoparticles for Biomedical Applications: Mimicking the Workhorses of Our Body.
Hock N, Racaniello GF, Aspinall S, Denora N, Khutoryanskiy VV, Bernkop-Schnürch A. Hock N, et al. Adv Sci (Weinh). 2022 Jan;9(1):e2102451. doi: 10.1002/advs.202102451. Epub 2021 Nov 12. Adv Sci (Weinh). 2022. PMID: 34773391 Free PMC article. Review. - Drug delivery systems, CNS protection, and the blood brain barrier.
Upadhyay RK. Upadhyay RK. Biomed Res Int. 2014;2014:869269. doi: 10.1155/2014/869269. Epub 2014 Jul 20. Biomed Res Int. 2014. PMID: 25136634 Free PMC article. Review. - Thiolated Chitosans: A Multi-talented Class of Polymers for Various Applications.
Federer C, Kurpiers M, Bernkop-Schnürch A. Federer C, et al. Biomacromolecules. 2021 Jan 11;22(1):24-56. doi: 10.1021/acs.biomac.0c00663. Epub 2020 Jul 9. Biomacromolecules. 2021. PMID: 32567846 Free PMC article. Review.
References
- Yadava PK. Nucleic acid therapeutics: current targets for antisense oligonucleotides and ribozymes. Molecular Biology Today. 2000:1–16.
- Stein CA. Two problems in antisense biotechnology: in vitro delivery and the design of antisense experiments. Biochimica et Biophysica Acta. 1999;1489(1):45–52. - PubMed
- Israel ZH, Domb AJ. Polymers in gene therapy: antisense delivery systems. Polym Adv Technol. 1998;9(10–11):799–805.
- Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55(3):329–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous