The dynamic nature of autophagy in cancer - PubMed (original) (raw)
Review
The dynamic nature of autophagy in cancer
Alec C Kimmelman. Genes Dev. 2011.
Abstract
Macroautophagy (referred to hereafter as autophagy) is a highly regulated cellular process that serves to remove damaged proteins and organelles from the cell. Autophagy contributes to an array of normal and pathological processes, and has recently emerged as a key regulator of multiple aspects of cancer biology. The role of autophagy in cancer is complex and is likely dependent on tumor type, stage, and genetic context. This complexity is illustrated by the identification of settings where autophagy acts potently to either promote or inhibit tumorigenesis. In this review, I discuss the underlying basis for these opposing functions and propose a model suggesting a dynamic role for autophagy in malignancy. Collectively, the data point to autophagy as serving as a barrier to limit tumor initiation. Once neoplastic lesions are established, it appears that adaptive changes occur that now result in positive roles for autophagy in malignant progression and in subsequent tumor maintenance. Remarkably, constitutive activation of autophagy is critical for continued growth of some tumors, serving to both reduce oxidative stress and provide key intermediates to sustain cell metabolism. Autophagy is also induced in response to cancer therapies where it can function as a survival mechanism that limits drug efficacy. These findings have inspired significant interest in applying anti-autophagy therapies as an entirely new approach to cancer treatment. It is now apparent that aberrant control of autophagy is among the key hallmarks of cancer. While much needs to be learned about the regulation and context-dependent biological functions of autophagy, it seems clear that modulation of this process will be an attractive avenue for future cancer therapeutic approaches.
Figures
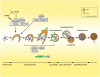
Figure 1.
The process and regulation of autophagy. The stages of autophagy (initiation, elongation, closure, maturation, and degradation) are depicted. Cargo is sequestered in a double-membrane vesicle that eventually forms the autophagosome. This fuses to the lysosome (autolysosome), where the cargo is degraded by lysosomal enzymes and degradation products are recycled back into the cytosol by lysosomal permeases. mTOR is a key regulator of autophagy in response to changes in nutrient availability. During nutrient-replete conditions, mTOR is activated and autophagy is inhibited through repression of ULK1/2 (the mammalian homologs of ATG1). Upon nutrient depletion, ULK1/2 is activated and can promote autophagy initiation. ULK is also activated in states of low energy (increased AMP/ATP ratio) by phosphorylation by AMPK as well as repression of mTORC1. Also critical to autophagy initiation is the production of phosphatidylinositol-3-phosphate (PI3P) by the class III PI3K Vps34, which is in a complex with ATG6/Beclin1 and p150 (Vps15). This complex is an additional level of regulation and, depending on the particular proteins bound, can activate or repress Vps34 activity. The ATG5–ATG12 complex as well as LC3 conjugated to PE, known as LC3-II, act downstream from Vps34 and ULK1/2 and, along with other proteins, have roles in autophagosome membrane elongation. LC3 acts downstream from the ATG5–ATG12 system and is present on the outer and inner surfaces of the autophagosome (depicted as a green oval). LC3 is a commonly used marker to monitor autophagosomes.
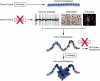
Figure 2.
Proposed model of the role of autophagy in cancer development and progression. In normal tissue, autophagy performs homeostatic functions such as organelle and protein quality control. If autophagy is suppressed in tissues, normal homeostasis is disrupted. Consequences of this include increased inflammation, genomic instability, and aneuploidy. Together, these changes can promote tumor initiation and lead to early tumorigeneisis (the schematic depicts an early lesion with increased nuclear atypia and loss of cell polarity, but an intact basement membrane in tan). However, if autophagy continues to be suppressed, then tumor progression will not proceed. Alternatively, if autophagy is activated at this stage, cells can keep up with their metabolic demand as well as regulate oxidative stress. This allows progression to more advanced malignancy (tumor breaks through the basement membrane) as well as continued tumor growth. Genomic instability is depicted as a recurrence plot from array comparative genomic hybridization of a pancreatic cancer cell line showing multiple high-amplitude genomic changes. Aneuploidy is shown by a spectral karyotype of a tumor cell line demonstrating aneuplody and chromosomal rearrangements.
Similar articles
- Autophagy and cancer cell metabolism.
Lozy F, Karantza V. Lozy F, et al. Semin Cell Dev Biol. 2012 Jun;23(4):395-401. doi: 10.1016/j.semcdb.2012.01.005. Epub 2012 Jan 18. Semin Cell Dev Biol. 2012. PMID: 22281437 Free PMC article. Review. - The multifaceted role of autophagy in tumor evasion from immune surveillance.
Janji B, Viry E, Moussay E, Paggetti J, Arakelian T, Mgrditchian T, Messai Y, Noman MZ, Van Moer K, Hasmim M, Mami-Chouaib F, Berchem G, Chouaib S. Janji B, et al. Oncotarget. 2016 Apr 5;7(14):17591-607. doi: 10.18632/oncotarget.7540. Oncotarget. 2016. PMID: 26910842 Free PMC article. Review. - Autophagy: an emerging target for cancer therapy.
Høyer-Hansen M, Jäättelä M. Høyer-Hansen M, et al. Autophagy. 2008 Jul;4(5):574-80. doi: 10.4161/auto.5921. Epub 2008 Mar 17. Autophagy. 2008. PMID: 18362515 Review. - Autophagy in malignant transformation and cancer progression.
Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, Kimmelman A, Kumar S, Levine B, Maiuri MC, Martin SJ, Penninger J, Piacentini M, Rubinsztein DC, Simon HU, Simonsen A, Thorburn AM, Velasco G, Ryan KM, Kroemer G. Galluzzi L, et al. EMBO J. 2015 Apr 1;34(7):856-80. doi: 10.15252/embj.201490784. Epub 2015 Feb 23. EMBO J. 2015. PMID: 25712477 Free PMC article. Review. - The Evolving, Multifaceted Roles of Autophagy in Cancer.
Liu J, Debnath J. Liu J, et al. Adv Cancer Res. 2016;130:1-53. doi: 10.1016/bs.acr.2016.01.005. Epub 2016 Feb 23. Adv Cancer Res. 2016. PMID: 27037750 Review.
Cited by
- Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis.
Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, Snyder E, Santanam U, Dipaola RS, Jacks T, Rabinowitz JD, White E. Guo JY, et al. Genes Dev. 2013 Jul 1;27(13):1447-61. doi: 10.1101/gad.219642.113. Genes Dev. 2013. PMID: 23824538 Free PMC article. - The Controversial Role of Autophagy in Ewing Sarcoma Pathogenesis-Current Treatment Options.
Koustas E, Sarantis P, Karamouzis MV, Vielh P, Theocharis S. Koustas E, et al. Biomolecules. 2021 Feb 26;11(3):355. doi: 10.3390/biom11030355. Biomolecules. 2021. PMID: 33652741 Free PMC article. Review. - Autophagy is a protective mechanism for human melanoma cells under acidic stress.
Marino ML, Pellegrini P, Di Lernia G, Djavaheri-Mergny M, Brnjic S, Zhang X, Hägg M, Linder S, Fais S, Codogno P, De Milito A. Marino ML, et al. J Biol Chem. 2012 Aug 31;287(36):30664-76. doi: 10.1074/jbc.M112.339127. Epub 2012 Jul 3. J Biol Chem. 2012. PMID: 22761435 Free PMC article. - HMGB1 is a therapeutic target for leukemia.
Yu Y, Xie M, Kang R, Livesey KM, Cao L, Tang D. Yu Y, et al. Am J Blood Res. 2012;2(1):36-43. Epub 2012 Jan 1. Am J Blood Res. 2012. PMID: 22432086 Free PMC article. - The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis.
Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. Roczniak-Ferguson A, et al. Sci Signal. 2012 Jun 12;5(228):ra42. doi: 10.1126/scisignal.2002790. Sci Signal. 2012. PMID: 22692423 Free PMC article.
References
- Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A 2008. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res 68: 1485–1494 - PubMed
- Apetoh L, Ghiringhelli F, Tesniere A, Criollo A, Ortiz C, Lidereau R, Mariette C, Chaput N, Mira JP, Delaloge S, et al. 2007. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol Rev 220: 47–59 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources