Versatile and efficient genome editing in human cells by combining zinc-finger nucleases with adeno-associated viral vectors - PubMed (original) (raw)
Versatile and efficient genome editing in human cells by combining zinc-finger nucleases with adeno-associated viral vectors
Eva-Maria Händel et al. Hum Gene Ther. 2012 Mar.
Abstract
Zinc-finger nucleases (ZFNs) have become a valuable tool for targeted genome engineering. Based on the enzyme's ability to create a site-specific DNA double-strand break, ZFNs promote genome editing by activating the cellular DNA damage response, including homology-directed repair (HDR) and nonhomologous end-joining. The goal of this study was (i) to demonstrate the versatility of combining the ZFN technology with a vector platform based on adeno-associated virus (AAV), and (ii) to assess the toxicity evoked by this platform. To this end, human cell lines that harbor enhanced green fluorescence protein (EGFP) reporters were generated to easily quantify the frequencies of gene deletion, gene disruption, and gene correction. We demonstrated that ZFN-encoding AAV expression vectors can be employed to induce large chromosomal deletions or to disrupt genes in up to 32% of transduced cells. In combination with AAV vectors that served as HDR donors, the AAV-ZFN platform was utilized to correct a mutation in EGFP in up to 6% of cells. Genome editing on the DNA level was confirmed by genotyping. Although cell cycle profiling revealed a modest G2/M arrest at high AAV-ZFN vector doses, platform-induced apoptosis could not be detected. In conclusion, the combined AAV-ZFN vector technology is a useful tool to edit the human genome with high efficiency. Because AAV vectors can transduce many cell types relevant for gene therapy, the ex vivo and in vivo delivery of ZFNs via AAV vectors will be of great interest for the treatment of inherited disorders.
Figures
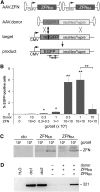
FIG. 1.
Adeno-associated virus (AAV)-mediated genome editing by nonhomologous end-joining (NHEJ). (A) Targeted deletion of lentiviral provirus. U2OS.693 cells carry an integrated copy of a destabilized enhanced green fluorescence protein (dsEGFP) expression cassette that was introduced by lentiviral transduction (target). The lentiviral long terminal repeats harbor a target site for zinc-finger nuclease (ZFN) pair EB/BA (fat arrows). Expression of both ZFN subunits (ZFNEB and ZFNBA) upon infection of cells with 3×103 (gray) or 105 genome copies (gc)/cell (black) of the respective AAV vectors leads to excision of the expression cassette (product). The graph shows the percentage of EGFP-negative cells, as assessed by flow cytometry; * indicates statistically significant increase in EGFP-negative cells as compared to cells infected with AAV-ZFNEB alone (p<0.05). (B) Polymerase chain reaction (PCR)-based molecular characterization. Genomic DNA of single EGFP-negative clones (lanes 1–8) was extracted and used as a template to amplify a 1-kb fragment contained in the provirus. The positions of the primers are indicated as arrows in (A). Amplification of a 150-bp fragment in the PTBP2 gene served as a control. NT, nontransduced U2OS.693 cells; U, U2OS parental cell line; H2O, PCR control. (C) Targeted gene disruption. U2OS.693 were infected with 103, 104, or 105 gc/cell of AAV vectors that express ZFNs targeting either position 292 or 502 (fat arrows) of the EGFP reading frame, respectively. Error-prone repair of the DNA double-strand break leads to disruption of the coding sequence (product). The graph shows the fraction of EGFP-negative cells, as assayed 5 days post-infection by flow cytometry; * indicates statistically significant increase in EGFP-negative cells, as compared to cells infected with a control vector expressing a nonfunctional nuclease (p<0.02). (D) T7 endonuclease I (T7E1)-based genotyping. Genomic DNA of cells transduced with E502-specific ZFN expression vectors was used as a template to amplify a 544-bp fragment containing the target site. The amplicon was subjected to digestion with mismatch-sensitive T7E1 to verify the presence of insertions/deletions at E502. The position of the expected 446-bp band is indicated. (E) PCR strategy to detect AAV-ZFN integration into E502 site in antisense orientation. The positions of the PCR primers to detect the 5′- and 3′-junctions, respectively, are shown. (F) Qualitative assessment. Genomic DNA of U2OS.693 cells transduced with E502-specific ZFN expression vectors was subjected to PCR analysis. DNA from nontransduced (NT) cells served as a control. The position of the expected ∼800-bp band is indicated. H2O, PCR control.
FIG. 2.
AAV-mediated genome editing by homology-directed repair (HDR). (A) Schematic of gene correction in U2OS.893 cells. The target locus consists of a mutated EGFP (mGFP) gene under control of a cytomegalovirus (CMV) promoter, followed by an ires-NeoR-wpre cassette. The mutation in mGFP is based on a 43-bp insertion that includes three in-frame stop codons and a recognition site for ZFN pair EB/BA (fat arrow). The AAV donor vector contains a 5′-truncated EGFP gene (∂GFP) followed by the ires-NeoR-wpre cassette. HDR (indicated by two crosses) between target locus and AAV donor leads to expression of a functional EGFP gene (product). (B) AAV-ZFN mediated gene targeting. U2OS.893 cells were transduced with the indicated vector dose of AAV donor and ZFN expression vectors, and assessed by flow cytometry 6 days post-transduction. The graph displays the average percentage of EGFP-positive cells; * and ** indicate statistically significant increase in EGFP-positive cells as compared to mock infected cells (p<0.01 and p<0.002, respectively). (C) AAV-mediated ZFN expression. U2OS cells were transduced separately with AAV-ZFNEB, AAV-ZFNBA, or a control vector at 104 or 105 gc/cell, harvested after 72 hr, and ZFN expression levels detected using an HA tag specific antibody. (D) Genotyping. Genomic DNA of U2OS.696 cells infected with 104 gc/cell of AAV donor or AAV-ZFN, as indicated, was isolated 30 days post-transduction. The DNA was subjected to nested PCR analysis (primer positions shown in panel A) to detect the corrected target locus; 104 and 105 copies of an EGFP plasmid served as positive controls (cto1, cto2).
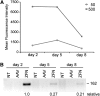
FIG. 3.
AAV expression kinetics. (A) Protein expression. U2OS cells were transduced with an AAV-EGFP vector at 50 or 500 gc/cell, respectively, and subjected to flow cytometric analysis at the indicated time points. The graph displays mean fluorescence intensity measured in channel FL-1. (B) RNA expression. U2OS cells were transduced with AAV-ZFNEB and AAV-ZFNBA at 105 gc/cell. RT-PCR amplifying a fragment contained in the ZFN _Fok_I domain was performed on RNA extracted from transduced cells at indicated time points. The position of the expected 162-bp band is indicated. Numbers below the gel indicate signal intensities relative to day 2. NT, nontransduced cells; AAV, cells transduced with an AAV control vector.
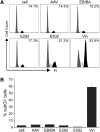
FIG. 4.
Platform-associated toxicity. U2OS.693 cells were infected with 105 gc/cell of AAV-ZFN vectors, as indicated, fixed in ethanol 4 days after infection, stained with propidium iodide (PI) and analyzed by flow cytometry. Cells treated with 100 nM vinblastine (Vin) were assayed after 1 day and served as a control. (A) Cell cycle profile. The plots show the respective cell populations in G1 (gray), S (white), and G2/M phase (black). The average percentage of cells in G2/M phase is indicated. (B) Apoptosis. The graph displays the average fraction of cells in the subG1 population at day 4 post-transduction.
Similar articles
- The nontoxic cell cycle modulator indirubin augments transduction of adeno-associated viral vectors and zinc-finger nuclease-mediated gene targeting.
Rahman SH, Bobis-Wozowicz S, Chatterjee D, Gellhaus K, Pars K, Heilbronn R, Jacobs R, Cathomen T. Rahman SH, et al. Hum Gene Ther. 2013 Jan;24(1):67-77. doi: 10.1089/hum.2012.168. Epub 2012 Dec 10. Hum Gene Ther. 2013. PMID: 23072634 Free PMC article. - Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs.
Ellis BL, Hirsch ML, Porter SN, Samulski RJ, Porteus MH. Ellis BL, et al. Gene Ther. 2013 Jan;20(1):35-42. doi: 10.1038/gt.2011.211. Epub 2012 Jan 19. Gene Ther. 2013. PMID: 22257934 Free PMC article. - Codon swapping of zinc finger nucleases confers expression in primary cells and in vivo from a single lentiviral vector.
Abarrategui-Pontes C, Créneguy A, Thinard R, Fine EJ, Thepenier V, Fournier le RL, Cradick TJ, Bao G, Tesson L, Podevin G, Anegon I, Nguyen TH. Abarrategui-Pontes C, et al. Curr Gene Ther. 2014;14(5):365-76. doi: 10.2174/156652321405140926161748. Curr Gene Ther. 2014. PMID: 25687502 - AAV Vectorization of DSB-mediated Gene Editing Technologies.
Moser RJ, Hirsch ML. Moser RJ, et al. Curr Gene Ther. 2016;16(3):207-19. doi: 10.2174/1566523216666160602213738. Curr Gene Ther. 2016. PMID: 27280971 Review. - Basic and Clinical Application of Adeno-Associated Virus-Mediated Genome Editing.
He X, Xie H, Liu X, Gu F. He X, et al. Hum Gene Ther. 2019 Jun;30(6):673-681. doi: 10.1089/hum.2018.190. Epub 2019 Feb 28. Hum Gene Ther. 2019. PMID: 30588843 Review.
Cited by
- Combinatorial anti-HIV gene therapy: using a multipronged approach to reach beyond HAART.
Peterson CW, Younan P, Jerome KR, Kiem HP. Peterson CW, et al. Gene Ther. 2013 Jul;20(7):695-702. doi: 10.1038/gt.2012.98. Epub 2013 Jan 31. Gene Ther. 2013. PMID: 23364313 Free PMC article. Review. - Improving Homology-Directed Repair in Genome Editing Experiments by Influencing the Cell Cycle.
Smirnikhina SA, Zaynitdinova MI, Sergeeva VA, Lavrov AV. Smirnikhina SA, et al. Int J Mol Sci. 2022 May 26;23(11):5992. doi: 10.3390/ijms23115992. Int J Mol Sci. 2022. PMID: 35682671 Free PMC article. Review. - Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells.
Liu J, Gaj T, Yang Y, Wang N, Shui S, Kim S, Kanchiswamy CN, Kim JS, Barbas CF 3rd. Liu J, et al. Nat Protoc. 2015 Nov;10(11):1842-59. doi: 10.1038/nprot.2015.117. Epub 2015 Oct 22. Nat Protoc. 2015. PMID: 26492140 - Genome-editing Technologies for Gene and Cell Therapy.
Maeder ML, Gersbach CA. Maeder ML, et al. Mol Ther. 2016 Mar;24(3):430-46. doi: 10.1038/mt.2016.10. Epub 2016 Jan 12. Mol Ther. 2016. PMID: 26755333 Free PMC article. Review. - Targeted multi-epitope switching enables straightforward positive/negative selection of CAR T cells.
Mosti L, Langner LM, Chmielewski KO, Arbuthnot P, Alzubi J, Cathomen T. Mosti L, et al. Gene Ther. 2021 Sep;28(9):602-612. doi: 10.1038/s41434-021-00220-6. Epub 2021 Feb 1. Gene Ther. 2021. PMID: 33526841 Free PMC article.
References
- Alwin S. Gere M.B. Guhl E., et al. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. - PubMed
- Benabdallah B.F. Allard E. Yao S., et al. Targeted gene addition to human mesenchymal stromal cells as a cell-based plasma-soluble protein delivery platform. Cytotherapy. 2010;12:394–399. - PubMed
- Chamberlain J.R. Schwarze U. Wang P.R., et al. Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science. 2004;303:1198–1201. - PubMed
- Cornu T.I. Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources