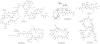A mass spectrometry-guided genome mining approach for natural product peptidogenomics - PubMed (original) (raw)
A mass spectrometry-guided genome mining approach for natural product peptidogenomics
Roland D Kersten et al. Nat Chem Biol. 2011.
Abstract
Peptide natural products show broad biological properties and are commonly produced by orthogonal ribosomal and nonribosomal pathways in prokaryotes and eukaryotes. To harvest this large and diverse resource of bioactive molecules, we introduce here natural product peptidogenomics (NPP), a new MS-guided genome-mining method that connects the chemotypes of peptide natural products to their biosynthetic gene clusters by iteratively matching de novo tandem MS (MS(n)) structures to genomics-based structures following biosynthetic logic. In this study, we show that NPP enabled the rapid characterization of over ten chemically diverse ribosomal and nonribosomal peptide natural products of previously unidentified composition from Streptomycete bacteria as a proof of concept to begin automating the genome-mining process. We show the identification of lantipeptides, lasso peptides, linardins, formylated peptides and lipopeptides, many of which are from well-characterized model Streptomycetes, highlighting the power of NPP in the discovery of new peptide natural products from even intensely studied organisms.
Figures
Figure 1
Structural diversity of peptide natural products.
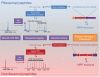
Figure 2. General workflow of Natural Product Peptidogenomics (NPP)
NPP can be applied to characterize both ribosomal and nonribosomal peptide natural products in their genotype and chemotype from genome-sequenced organisms. Two proof of concept NPP experiments are outlined: Ribosomal peptides (RNPs) or nonribosomal peptides (NRPs) and their respective biosynthetic gene cluster can be characterized from a Streptomyces extract by MALDI-TOF MS detection, MSn sequence tagging and PNP genome mining. The iterative approach in matching MSn data to genomics-derived peptide structures is shown by dashed arrows. See Figures 3 and 4, respectively, for detailed NPP analysis of ribosomal and nonribosomal peptides.
Figure 3. Peptidogenomic connection of a ribosomal peptide (RNP) chemotype with its biosynthetic genes (genotype) via sequence tagging and genome mining – Characterization of class III lantipeptide AmfS from Streptomyces griseus IFO 13350
Iterative aspects in connecting MS data of the peptide chemotype to the genotype are highlighted in blue and in dashed arrows. Steps are as follows: (A) Detection of putative peptide mass signals by MALDI-TOF MS or Imaging MS, (B) determination of molecular weight, (C) MSn fragmentation (CID), (D) assignment of charge states, (E) identification of mass shifts, (F) substitution of proteinogenic mass shifts (Supplementary Table 5), (G) substitution of nonproteinogenic mass shifts with putative RNP monomers (Supplementary Table 6), (H) MSn sequence tag processing of putative biosynthetic or MS gas-phase modifications, (I) MSn sequence tag processing of sequence tag direction, (J) search in 6-frame translation of target genome, (K) identification of candidate precursor peptide via RNP biosynthetic rationale, (L) verification of precursor peptide sequence, (M) prediction of core peptide sequence based on observed mass and putative PTM mass shifts, (N) verification of core peptide sequence and mass, (O) prediction of biosynthetic gene cluster, (P) verification of putative PTMs, (Q) RNP classification, (R) structure prediction based on RNP class and MSn data, and (S) structure verification by MSn data.
Figure 4. Peptidogenomic connection of a nonribosomal peptide (NRP) chemotype with its biosynthetic genes (genotype) via sequence tagging and genome mining – Characterization of the lipopeptide stendomycin complex from Streptomyces hygroscopicus ATCC 53653
Iterative aspects in connecting MSn data of the peptide chemotype to the genotype are highlighted in blue and in dashed arrows. Steps are as follows: (A) Detection of putative peptide mass signals by MALDI-TOF MS or Imaging-MS, (B) determination of molecular weight, (C) MSn fragmentation (CID), (D) Assignment of charge states, (E) identification of mass shifts, (F) substitution of proteinogenic mass shifts (Supplementary Table 5), (G) substitution of nonproteinogenic mass shifts with putative NRP monomers (Supplementary Table 7), (H) MSn sequence tag processing of putative biosynthetic and MS gas-phase modifications, (I) MSn sequence tag processing of sequence tag direction, (J) search predicted NRP sequences from target genome with all search tags (NP.searcher/antiSMASH), (K) biosynthetic gene cluster analysis, (L) verification of predicted NRPS assembly-line, (M) NRP structure prediction, (N) verification of predicted NRP structure, (O) full structure elucidation based on MSn and NMR data, and (P) verification of NRP structure. Abbreviations: Dha – dehydroalanine; Dhb – dehydrobutyrine; HseL – homoserine lactone; Orn – ornithine.
Similar articles
- Automated genome mining of ribosomal peptide natural products.
Mohimani H, Kersten RD, Liu WT, Wang M, Purvine SO, Wu S, Brewer HM, Pasa-Tolic L, Bandeira N, Moore BS, Pevzner PA, Dorrestein PC. Mohimani H, et al. ACS Chem Biol. 2014 Jul 18;9(7):1545-51. doi: 10.1021/cb500199h. Epub 2014 May 23. ACS Chem Biol. 2014. PMID: 24802639 Free PMC article. - Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products.
Medema MH, Paalvast Y, Nguyen DD, Melnik A, Dorrestein PC, Takano E, Breitling R. Medema MH, et al. PLoS Comput Biol. 2014 Sep 4;10(9):e1003822. doi: 10.1371/journal.pcbi.1003822. eCollection 2014 Sep. PLoS Comput Biol. 2014. PMID: 25188327 Free PMC article. - Dereplication, sequencing and identification of peptidic natural products: from genome mining to peptidogenomics to spectral networks.
Mohimani H, Pevzner PA. Mohimani H, et al. Nat Prod Rep. 2016 Jan;33(1):73-86. doi: 10.1039/c5np00050e. Nat Prod Rep. 2016. PMID: 26497201 Free PMC article. Review. - Prospecting genomes for lasso peptides.
Maksimov MO, Link AJ. Maksimov MO, et al. J Ind Microbiol Biotechnol. 2014 Feb;41(2):333-44. doi: 10.1007/s10295-013-1357-4. Epub 2013 Oct 19. J Ind Microbiol Biotechnol. 2014. PMID: 24142336 Review. - A new genome-mining tool redefines the lasso peptide biosynthetic landscape.
Tietz JI, Schwalen CJ, Patel PS, Maxson T, Blair PM, Tai HC, Zakai UI, Mitchell DA. Tietz JI, et al. Nat Chem Biol. 2017 May;13(5):470-478. doi: 10.1038/nchembio.2319. Epub 2017 Feb 28. Nat Chem Biol. 2017. PMID: 28244986 Free PMC article.
Cited by
- A metabologenomics strategy for rapid discovery of polyketides derived from modular polyketide synthases.
Liu RZ, Zhang Z, Li M, Zhang L. Liu RZ, et al. Chem Sci. 2024 Nov 4. doi: 10.1039/d4sc04174g. Online ahead of print. Chem Sci. 2024. PMID: 39568943 Free PMC article. - Advances in lasso peptide discovery, biosynthesis, and function.
Barrett SE, Mitchell DA. Barrett SE, et al. Trends Genet. 2024 Nov;40(11):950-968. doi: 10.1016/j.tig.2024.08.002. Epub 2024 Aug 31. Trends Genet. 2024. PMID: 39218755 Review. - Dehydroamino acid residues in bioactive natural products.
Wang S, Wu K, Tang YJ, Deng H. Wang S, et al. Nat Prod Rep. 2024 Feb 21;41(2):273-297. doi: 10.1039/d3np00041a. Nat Prod Rep. 2024. PMID: 37942836 Free PMC article. Review. - Peptidomics Methods Applied to the Study of Flower Development.
Álvarez-Urdiola R, Borràs E, Valverde F, Matus JT, Sabidó E, Riechmann JL. Álvarez-Urdiola R, et al. Methods Mol Biol. 2023;2686:509-536. doi: 10.1007/978-1-0716-3299-4_24. Methods Mol Biol. 2023. PMID: 37540375 - sORFPred: A Method Based on Comprehensive Features and Ensemble Learning to Predict the sORFs in Plant LncRNAs.
Chen Z, Meng J, Zhao S, Yin C, Luan Y. Chen Z, et al. Interdiscip Sci. 2023 Jun;15(2):189-201. doi: 10.1007/s12539-023-00552-4. Epub 2023 Jan 27. Interdiscip Sci. 2023. PMID: 36705893
References
- Daffre S, et al. Bioactive natural peptides. In: Rahman AU, editor. Studies in Natural Products Chemistry. 1st edn. Vol. 35. Elsevier; Oxford: 2008. pp. 597–691.
- Donadio S, Monciardini P, Sosio M. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat. Prod. Rep. 2007;24:1073–1109. - PubMed
- Ganz T. Defensins and host defense. Science. 1999;286:420–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 CA044848/CA/NCI NIH HHS/United States
- R01 GM086283/GM/NIGMS NIH HHS/United States
- R01 GM085770-04/GM/NIGMS NIH HHS/United States
- R37 CA044848-25/CA/NCI NIH HHS/United States
- R01 GM085770/GM/NIGMS NIH HHS/United States
- GM086283/GM/NIGMS NIH HHS/United States
- R01 GM086283-03/GM/NIGMS NIH HHS/United States
- GM085770/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous