The architecture of long-range haplotypes shared within and across populations - PubMed (original) (raw)
The architecture of long-range haplotypes shared within and across populations
Alexander Gusev et al. Mol Biol Evol. 2012 Feb.
Abstract
Homologous long segments along the genomes of close or remote relatives that are identical by descent (IBD) from a common ancestor provide clues for recent events in human genetics. We set out to extensively map such IBD segments in large cohorts and investigate their distribution within and across different populations. We report analysis of several data sets, demonstrating that IBD is more common than expected by naïve models of population genetics. We show that the frequency of IBD pairs is population dependent and can be used to cluster individuals into populations, detect a homogeneous subpopulation within a larger cohort, and infer bottleneck events in such a subpopulation. Specifically, we show that Ashkenazi Jewish individuals are all connected through transitive remote family ties evident by sharing of 50 cM IBD to a publicly available data set of less than 400 individuals. We further expose regions where long-range haplotypes are shared significantly more often than elsewhere in the genome, observed across multiple populations, and enriched for common long structural variation. These are inconsistent with recent relatedness and suggest ancient common ancestry, with limited recombination between haplotypes.
Figures
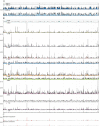
FIG. 1.
Manhattan-style plots of IBD segment sharing in worldwide populations. Fraction of pairs of individuals IBD, on the y axis, at a locus shown as a function of the genomic position at the locus (A) within Ashkenazi/European cohorts, (B) within HapMap cohorts, and (C) between HapMap continents/populations (scale not consistent with A, B). Panel c highlights enriched regions, consistent with intrapopulation sharing. Within populations, the normalization factor was equal to the number of unique pairs; between populations, the normalization factor was the product of the respective cohort sizes.
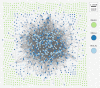
FIG. 2.
Graph plot of IBD sharing in HapMap populations and resultant clusters. Nodes denote individuals, color-coded by cohort, and edges represent normalized genome-wide IBD sharing. (A) Initial clusters from unfiltered sharing—{GIH},{LWK},{JPT,CHD,CHB},{CEU,TSI} segregate. (b) Final clusters after cross-cluster edges have been iteratively removed—{TSI},{CEU} newly segregated.
FIG. 3.
Graph plot of IBD sharing between samples of Ashkenazi (blue/dark) and European (green/light) origin. Each colored vertex represents a sample from the respective population, edges represent IBD sharing between incident individuals, and edge width represents total amount of sharing genomewide. Ashkenazi samples form “giant connected component” and no edges longer than 100 cM to the European population.
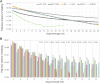
FIG. 4.
Relationship between segment length and amount of sharing in real and simulated data. We compute the expected number of IBD segments shared within each population (y axis, logarithmic scale) for the discrete segment length range of 3 to 30 cM (x axis). (A) AJ and EU populations shown with dot and line, solid lines show simulated coalescent data rawn from a Wright–Fisher model (WF—dark/light gray) and a bottleneck model (BN—highlight). (B) HapMap populations shown in solid colors. Y-intercept correlates to ancestral population size, decay loosely correlates to population growth. For both figures, only data points at which sharing is more than 1 in a 1,000 pairs of individuals (varies by population) are shown.
Similar articles
- No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews.
Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, Tzur S, Sahakyan H, Bahmanimehr A, Yepiskoposyan L, Tambets K, Khusnutdinova EK, Kushniarevich A, Balanovsky O, Balanovsky E, Kovacevic L, Marjanovic D, Mihailov E, Kouvatsi A, Triantaphyllidis C, King RJ, Semino O, Torroni A, Hammer MF, Metspalu E, Skorecki K, Rosset S, Halperin E, Villems R, Rosenberg NA. Behar DM, et al. Hum Biol. 2013 Dec;85(6):859-900. doi: 10.3378/027.085.0604. Hum Biol. 2013. PMID: 25079123 - Length distributions of identity by descent reveal fine-scale demographic history.
Palamara PF, Lencz T, Darvasi A, Pe'er I. Palamara PF, et al. Am J Hum Genet. 2012 Nov 2;91(5):809-22. doi: 10.1016/j.ajhg.2012.08.030. Epub 2012 Oct 25. Am J Hum Genet. 2012. PMID: 23103233 Free PMC article. - The variance of identity-by-descent sharing in the Wright-Fisher model.
Carmi S, Palamara PF, Vacic V, Lencz T, Darvasi A, Pe'er I. Carmi S, et al. Genetics. 2013 Mar;193(3):911-28. doi: 10.1534/genetics.112.147215. Epub 2012 Dec 24. Genetics. 2013. PMID: 23267057 Free PMC article. - Identity by descent between distant relatives: detection and applications.
Browning SR, Browning BL. Browning SR, et al. Annu Rev Genet. 2012;46:617-33. doi: 10.1146/annurev-genet-110711-155534. Epub 2012 Sep 17. Annu Rev Genet. 2012. PMID: 22994355 Review. - Identity by descent: variation in meiosis, across genomes, and in populations.
Thompson EA. Thompson EA. Genetics. 2013 Jun;194(2):301-26. doi: 10.1534/genetics.112.148825. Genetics. 2013. PMID: 23733848 Free PMC article. Review.
Cited by
- Genetic contribution to multiple sclerosis risk among Ashkenazi Jews.
Khankhanian P, Matsushita T, Madireddy L, Lizée A, Din L, Moré JM, Gourraud PA, Hauser SL, Baranzini SE, Oksenberg JR. Khankhanian P, et al. BMC Med Genet. 2015 Jul 28;16:55. doi: 10.1186/s12881-015-0201-2. BMC Med Genet. 2015. PMID: 26212423 Free PMC article. - Using identity by descent estimation with dense genotype data to detect positive selection.
Han L, Abney M. Han L, et al. Eur J Hum Genet. 2013 Feb;21(2):205-11. doi: 10.1038/ejhg.2012.148. Epub 2012 Jul 11. Eur J Hum Genet. 2013. PMID: 22781100 Free PMC article. - Demographic history differences between Hispanics and Brazilians imprint haplotype features.
da Cruz PRS, Ananina G, Secolin R, Gil-da-Silva-Lopes VL, Lima CSP, de França PHC, Donatti A, Lourenço GJ, de Araujo TK, Simioni M, Lopes-Cendes I, Costa FF, de Melo MB. da Cruz PRS, et al. G3 (Bethesda). 2022 Jul 6;12(7):jkac111. doi: 10.1093/g3journal/jkac111. G3 (Bethesda). 2022. PMID: 35511163 Free PMC article. - Linkage disequilibrium and within-breed genetic diversity in Iranian Zandi sheep.
Ghoreishifar SM, Moradi-Shahrbabak H, Parna N, Davoudi P, Khansefid M. Ghoreishifar SM, et al. Arch Anim Breed. 2019 Apr 2;62(1):143-151. doi: 10.5194/aab-62-143-2019. eCollection 2019. Arch Anim Breed. 2019. PMID: 31807624 Free PMC article. - Identifying recent adaptations in large-scale genomic data.
Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, Cabili M, Adegbola RA, Bamezai RN, Hill AV, Vannberg FO, Rinn JL; 1000 Genomes Project; Lander ES, Schaffner SF, Sabeti PC. Grossman SR, et al. Cell. 2013 Feb 14;152(4):703-13. doi: 10.1016/j.cell.2013.01.035. Cell. 2013. PMID: 23415221 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources