Organization and evolution of the cotG and cotH genes of Bacillus subtilis - PubMed (original) (raw)
Organization and evolution of the cotG and cotH genes of Bacillus subtilis
Rosa Giglio et al. J Bacteriol. 2011 Dec.
Abstract
The cotG and cotH genes of Bacillus subtilis encode two previously characterized spore coat proteins. The two genes are adjacent on the chromosome and divergently transcribed by σ(K), a sporulation-specific σ factor of the RNA polymerase. We report evidence that the cotH promoter maps 812 bp upstream of the beginning of its coding region and that the divergent cotG gene is entirely contained between the promoter and the coding part of cotH. A bioinformatic analysis of all entirely sequenced prokaryotic genomes showed that such chromosomal organization is not common in spore-forming bacilli. Indeed, CotG is present only in B. subtilis, B. amyloliquefaciens, and B. atrophaeus and in two Geobacillus strains. When present, cotG always encodes a modular protein composed of tandem repeats and is always close to but divergently transcribed with respect to cotH. Bioinformatic and phylogenic data suggest that such genomic organizations have a common evolutionary origin and that the modular structure of the extant cotG genes is the outcome of multiple rounds of gene elongation events of an ancestral minigene.
Figures
Fig. 1.
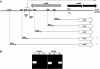
(A) Deletion analysis of the DNA region upstream of the cotH coding part. Numbers indicate positions on the DNA sequence, considering the first base of the translation start site as +1. The lacZ gene of E. coli is fused in frame to the cotH coding part as described by Baccigalupi et al. (2). β-gal, β-galactosidase. Thin, short arrows indicate the positions of the oligonucleotides used in the RT-PCR experiment corresponding to panel B, while thick gray and black arrows indicate the coding parts of cotG and cotH, respectively. The dashed arrow indicates the mRNA produced from the cotH promoter. (B) Agarose gel electrophoresis used to analyze the extension products of an RT-PCR experiment. Total RNA was extracted from sporulating cells 5 h after the onset of sporulation from a wild-type (PY79) strain. cDNA synthesis was primed with oligonucleotide H (panel A; Table 1), while the amplification reactions were primed with oligonucleotide pairs H/G22 and H/G24 (panel A; Table 1). Control experiments were performed using chromosomal DNA as a template (lanes 1 and 4) or mRNA without the addition of the RT enzyme (lanes 2 and 5). cDNA was used as a template in the reactions of lanes 3 and 6.
Fig. 2.
(A) Expression of a cotH::lacZ transcriptional fusion during sporulation in an otherwise wild-type (closed symbols) or gerE null mutant (open symbols) strain. Samples were collected at various times after the onset of sporulation. Enzyme activity is expressed in Miller units. Data are the means of three independent experiments, and error bars indicate the standard deviations. (B) Primer extension analysis of the cotH promoter region performed with total RNA extracted from sporulating cells 5 h after the onset of sporulation from a mutant that fails to make SigK (spoIVB::erm; lane 1), a gerE null mutant (gerE36; lane 3), and a wild-type strain (PY79; lane 2). Primer extension and sequencing reactions were primed with the synthetic oligonucleotide G25 (Table 1). (C) cotH promoter region. The transcription start site (“A”) is in bold and indicated as +1. The putative promoter sequences are indicated and the putative GerE boxes underlined. A comparison of the four GerE boxes with the GerE consensus is also reported.
Fig. 3.
(A) β-Galactosidase activity of strains carrying the cotH promoter and either 53 (squares) or 812 (circles) bp downstream of the transcription start site fused to the lacZ gene of E. coli. Gene fusions were inserted in an otherwise wild-type (open symbols) or gerE null mutant (closed symbols) strain. Samples were collected at various times after the onset of sporulation, and enzyme activity is expressed in Miller units. Data are the means of three independent experiments, and error bars indicate the standard deviations. (B) Construction of the deletion mutant. (C) Western blot of proteins extracted from sporulating cells or from purified spores of a wild-type strain and of an isogenic mutant carrying the deletion indicated in panel B. Proteins were fractionated on 15% polyacrylamide gels, electrotransferred to membranes, and reacted with anti-CotH antibody.
Fig. 4.
(A) Amino acid sequence of CotG of B. subtilis subsp. subtilis 168. The 19 modules present in the central part of the protein are indicated. The 7-amino-acid modules are in bold. (B) The 19 paralogous regions, 12 of 21 bp and 7 of 18 bp, encoding the protein modules.
Fig. 5.
Model for the origin and evolution of the cotG gene of Bacillus subtilis, considering the 21-bp module as the ancestral one.
Fig. 6.
(A) Amino acid modules of CotG of B. subtilis subsp. spizizenii W23, B. amyloliquefaciens FZB42, and B. atrophaeus 1942. (B) Amino acid sequence of CotG of Geobacillus WCH70. The three identical modules of 6 amino acids present in the central part of the protein are boxed. (C) Amino acid sequence of CotG of Geobacillus Y4.1MC1. The two 49 repeats present in the central part of the protein are boxed.
Similar articles
- CotG-Like Modular Proteins Are Common among Spore-Forming Bacilli.
Saggese A, Isticato R, Cangiano G, Ricca E, Baccigalupi L. Saggese A, et al. J Bacteriol. 2016 Apr 28;198(10):1513-20. doi: 10.1128/JB.00023-16. Print 2016 May 15. J Bacteriol. 2016. PMID: 26953338 Free PMC article. - Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat.
Zilhão R, Serrano M, Isticato R, Ricca E, Moran CP Jr, Henriques AO. Zilhão R, et al. J Bacteriol. 2004 Feb;186(4):1110-9. doi: 10.1128/JB.186.4.1110-1119.2004. J Bacteriol. 2004. PMID: 14762006 Free PMC article. - Bacillus subtilis spore coat assembly requires cotH gene expression.
Naclerio G, Baccigalupi L, Zilhao R, De Felice M, Ricca E. Naclerio G, et al. J Bacteriol. 1996 Aug;178(15):4375-80. doi: 10.1128/jb.178.15.4375-4380.1996. J Bacteriol. 1996. PMID: 8755863 Free PMC article. - Assembly and genetics of spore protective structures.
Takamatsu H, Watabe K. Takamatsu H, et al. Cell Mol Life Sci. 2002 Mar;59(3):434-44. doi: 10.1007/s00018-002-8436-4. Cell Mol Life Sci. 2002. PMID: 11964122 Free PMC article. Review. - [Identification and characterization of the outermost layer of Bacillus subtilis spores].
Imamura D. Imamura D. Yakugaku Zasshi. 2012;132(8):919-24. doi: 10.1248/yakushi.132.919. Yakugaku Zasshi. 2012. PMID: 22864350 Review. Japanese.
Cited by
- CotG controls spore surface formation in response to the temperature of growth in Bacillus subtilis.
Di Gregorio Barletta G, Vittoria M, Lanzilli M, Petrillo C, Ricca E, Isticato R. Di Gregorio Barletta G, et al. Environ Microbiol. 2022 Apr;24(4):2078-2088. doi: 10.1111/1462-2920.15960. Epub 2022 Mar 8. Environ Microbiol. 2022. PMID: 35254711 Free PMC article. - A protein phosphorylation module patterns the Bacillus subtilis spore outer coat.
Freitas C, Plannic J, Isticato R, Pelosi A, Zilhão R, Serrano M, Baccigalupi L, Ricca E, Elsholz AKW, Losick R, O Henriques A. Freitas C, et al. Mol Microbiol. 2020 Dec;114(6):934-951. doi: 10.1111/mmi.14562. Epub 2020 Sep 12. Mol Microbiol. 2020. PMID: 32592201 Free PMC article. - Bacillus subtilis Spore Surface Display of Haloalkane Dehalogenase DhaA.
Wang F, Song T, Jiang H, Pei C, Huang Q, Xi H. Wang F, et al. Curr Microbiol. 2019 Oct;76(10):1161-1167. doi: 10.1007/s00284-019-01723-7. Epub 2019 Jul 5. Curr Microbiol. 2019. PMID: 31278426 - Antagonistic role of CotG and CotH on spore germination and coat formation in Bacillus subtilis.
Saggese A, Scamardella V, Sirec T, Cangiano G, Isticato R, Pane F, Amoresano A, Ricca E, Baccigalupi L. Saggese A, et al. PLoS One. 2014 Aug 12;9(8):e104900. doi: 10.1371/journal.pone.0104900. eCollection 2014. PLoS One. 2014. PMID: 25115591 Free PMC article. - The Exosporium of Bacillus megaterium QM B1551 Is Permeable to the Red Fluorescence Protein of the Coral Discosoma sp.
Lanzilli M, Donadio G, Addevico R, Saggese A, Cangiano G, Baccigalupi L, Christie G, Ricca E, Isticato R. Lanzilli M, et al. Front Microbiol. 2016 Nov 4;7:1752. doi: 10.3389/fmicb.2016.01752. eCollection 2016. Front Microbiol. 2016. PMID: 27867376 Free PMC article.
References
- Baccigalupi L., et al. 2004. GerE-independent expression of cotH leads to CotC accumulation in the mother cell compartment during Bacillus subtilis sporulation. Microbiology 150:3441–3449 - PubMed
- Cutting S., Vander Horn P. B. 1990. Genetic analysis, p. 27–74 In Harwood C., Cutting S. (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom
- Driks A., Roels S., Beall B., Moran C. P., Jr., Losick R. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234–244 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases