Toward an understanding of the protein interaction network of the human liver - PubMed (original) (raw)
doi: 10.1038/msb.2011.67.
Keke Huo, Lixin Ma, Liujun Tang, Dong Li, Xiaobi Huang, Yanzhi Yuan, Chunhua Li, Wei Wang, Wei Guan, Hui Chen, Chaozhi Jin, Juncheng Wei, Wanqiao Zhang, Yongsheng Yang, Qiongming Liu, Ying Zhou, Cuili Zhang, Zhihao Wu, Wangxiang Xu, Ying Zhang, Tao Liu, Donghui Yu, Yaping Zhang, Liang Chen, Dewu Zhu, Xing Zhong, Lixin Kang, Xiang Gan, Xiaolan Yu, Qi Ma, Jing Yan, Li Zhou, Zhongyang Liu, Yunping Zhu, Tao Zhou, Fuchu He, Xiaoming Yang
Affiliations
- PMID: 21988832
- PMCID: PMC3261708
- DOI: 10.1038/msb.2011.67
Toward an understanding of the protein interaction network of the human liver
Jian Wang et al. Mol Syst Biol. 2011.
Erratum in
- Toward an understanding of the protein interaction network of the human liver.
Wang J, Huo K, Ma L, Tang L, Li D, Huang X, Yuan Y, Li C, Wang W, Guan W, Chen H, Jin C, Wei J, Zhang W, Yang Y, Liu Q, Zhou Y, Zhang C, Wu Z, Xu W, Zhang Y, Liu T, Yu D, Zhang Y, Chen L, Zhu D, Zhong X, Kang L, Gan X, Yu X, Ma Q, Yan J, Zhou L, Liu Z, Zhu Y, Zhou T, He F, Yang X. Wang J, et al. Mol Syst Biol. 2017 Dec 18;13(12):965. doi: 10.15252/msb.20178107. Mol Syst Biol. 2017. PMID: 29254952 Free PMC article.
Abstract
Proteome-scale protein interaction maps are available for many organisms, ranging from bacteria, yeast, worms and flies to humans. These maps provide substantial new insights into systems biology, disease research and drug discovery. However, only a small fraction of the total number of human protein-protein interactions has been identified. In this study, we map the interactions of an unbiased selection of 5026 human liver expression proteins by yeast two-hybrid technology and establish a human liver protein interaction network (HLPN) composed of 3484 interactions among 2582 proteins. The data set has a validation rate of over 72% as determined by three independent biochemical or cellular assays. The network includes metabolic enzymes and liver-specific, liver-phenotype and liver-disease proteins that are individually critical for the maintenance of liver functions. The liver enriched proteins had significantly different topological properties and increased our understanding of the functional relationships among proteins in a liver-specific manner. Our data represent the first comprehensive description of a HLPN, which could be a valuable tool for understanding the functioning of the protein interaction network of the human liver.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
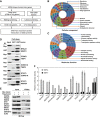
Construction of the proteome-scale HLPN. (A) Strategy of high-throughput Y2H screening. (B, C) Distribution of the GO categories of cellular component (CC) and molecular function (MF). From inside to outside, the rings represent all of the human proteins (16 022 with CC and 15 259 with MF annotations), human liver proteins (12 066 with CC and 10 863 with MF annotations), Y2H matrix proteins (4553 with CC and 4374 with MF annotations) and HLPN proteins (2342 with CC and 2252 with MF annotations). Each section reflects the percentage of proteins assigned to the given GO category. (D) GST pull-down assay. Bacteria-expressed GST or GST-tagged proteins were immobilized on glutathione-Sepharose 4B beads, and the beads were subsequently incubated with the Myc- or Flag-tagged proteins expressed in the HEK293T cell lysates. The proteins were detected using the indicated antibodies. (E) Co-IP assay. Flag- or Myc-tagged plasmids were transfected into HEK293T cells. Immunoprecipitations were performed using anti-Myc or anti-Flag antibodies and protein A/G-agarose. The lysates and immunoprecipitates were detected using the indicated antibodies. Myc-, GST- and Flag-fusions mean the fusion proteins with Myc, GST or Flag tags. (F) Luciferase reporter gene assays of SMAD3. Data are presented as mean values±s.d. (_n_=3). The results are representative of three independent experiments.
Figure 2
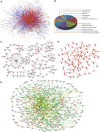
Network views of the HLPN. (A) Visualization of the HLPN. Yellow circles, selected Y2H proteins; red lines, interactions identified by library screening; blue lines, interactions identified by array screening; green lines, overlapping interactions of library and array screening; bold edges, interactions reported in the HPRD. (B) Distribution of the GO categories of biological processes for metabolic enzymes and their partners in the HLPN. The sections reflect the percentage of proteins assigned to the given GO category. (C) The ROS subnetwork. Blue circles, ROS proteins; red lines, data from the HPRD. (D) The liver-phenotype proteins network. Bold edges, interactions reported in the HPRD. (E) Liver-disease-associated proteins network. Red circles, HCC; cyan circles, cholangiocellular carcinoma; blue circles, liver fibrosis; yellow circles, other liver-disease proteins; gray circles, proteins without human liver disease annotation in the Library of Molecular Associations database (
http://www.medicalgenomics.org/databases/loma
); red lines: interactions among liver-disease proteins; yellow lines, interactions among non-liver-disease proteins; green lines, other interactions.
Figure 3
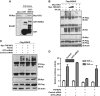
GIT2 is a negative regulator of the NF-κB signaling pathway. (A) Confirmation of the interaction between GIT2 and IKBKG by the GST pull-down assay. (B) GIT2 enhances the TNFAIP3-dependent deubiquitination of IKBKG. HEK293 cells were transfected with the indicated plasmids. Cell extracts were immunoprecipitated with anti-Flag antibody and detected by western blotting. (C) Knockdown of GIT2 by siRNA impairs the TNFAIP3-dependent deubiquitination of IKBKG. HEK293 cells were cotransfected with the indicated plasmids or the GIT2 siRNA. Cell extracts were immunoprecipitated with an anti-Flag antibody and detected by western blotting. (D) Knockdown of GIT2 impairs the TNFAIP3-mediated inhibition of NF-κB activity. HEK293 cells were cotransfected with the indicated plasmids. One day after transfection, the cells were stimulated with TNF-α for 6 h. Relative reporter activity was measured. Data are presented as mean values±s.d. (_n_=3). The results are representative of three independent experiments.
Similar articles
- Towards a proteome-scale map of the human protein-protein interaction network.
Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Rual JF, et al. Nature. 2005 Oct 20;437(7062):1173-8. doi: 10.1038/nature04209. Epub 2005 Sep 28. Nature. 2005. PMID: 16189514 - Interactome: gateway into systems biology.
Cusick ME, Klitgord N, Vidal M, Hill DE. Cusick ME, et al. Hum Mol Genet. 2005 Oct 15;14 Spec No. 2:R171-81. doi: 10.1093/hmg/ddi335. Epub 2005 Sep 14. Hum Mol Genet. 2005. PMID: 16162640 Review. - The Cartographers toolbox: building bigger and better human protein interaction networks.
Sanderson CM. Sanderson CM. Brief Funct Genomic Proteomic. 2009 Jan;8(1):1-11. doi: 10.1093/bfgp/elp003. Epub 2009 Mar 12. Brief Funct Genomic Proteomic. 2009. PMID: 19282470 Review. - Proteomics: The interaction map.
Baker M. Baker M. Nature. 2012 Apr 11;484(7393):271-5. doi: 10.1038/484271a. Nature. 2012. PMID: 22498631 No abstract available. - Analysis of protein-protein interactions using array-based yeast two-hybrid screens.
Rajagopala SV, Uetz P. Rajagopala SV, et al. Methods Mol Biol. 2009;548:223-45. doi: 10.1007/978-1-59745-540-4_13. Methods Mol Biol. 2009. PMID: 19521828
Cited by
- PAI-1-Dependent Inactivation of SMAD4-Modulated Junction and Adhesion Complex in Obese Endometrial Cancer.
Lin LL, Kost ER, Lin CL, Valente P, Wang CM, Kolonin MG, Daquinag AC, Tan X, Lucio N, Hung CN, Wang CP, Kirma NB, Huang TH. Lin LL, et al. Cell Rep. 2020 Oct 13;33(2):108253. doi: 10.1016/j.celrep.2020.108253. Cell Rep. 2020. PMID: 33053339 Free PMC article. - UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome.
Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, Osborne G, Bates GP, Glickman MH, Trost M, Knebel A, Marchesi F, Kurz T. Hjerpe R, et al. Cell. 2016 Aug 11;166(4):935-949. doi: 10.1016/j.cell.2016.07.001. Epub 2016 Jul 28. Cell. 2016. PMID: 27477512 Free PMC article. - ECR-MAPK regulation in liver early development.
Zhao XJ, Zhuo H. Zhao XJ, et al. Biomed Res Int. 2014;2014:850802. doi: 10.1155/2014/850802. Epub 2014 Dec 18. Biomed Res Int. 2014. PMID: 25580437 Free PMC article. - Inferring the Effects of Protein Variants on Protein-Protein Interactions with Interpretable Transformer Representations.
Liu Z, Qian W, Cai W, Song W, Wang W, Maharjan DT, Cheng W, Chen J, Wang H, Xu D, Lin GN. Liu Z, et al. Research (Wash D C). 2023 Sep 11;6:0219. doi: 10.34133/research.0219. eCollection 2023. Research (Wash D C). 2023. PMID: 37701056 Free PMC article. - Interaction of SMAC with a survivin-derived peptide alters essential cancer hallmarks: Tumor growth, inflammation, and immunosuppression.
Santhanam M, Kumar Pandey S, Shteinfer-Kuzmine A, Paul A, Abusiam N, Zalk R, Shoshan-Barmatz V. Santhanam M, et al. Mol Ther. 2024 Jun 5;32(6):1934-1955. doi: 10.1016/j.ymthe.2024.04.007. Epub 2024 Apr 5. Mol Ther. 2024. PMID: 38582961
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
- Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M (2008) Computing topological parameters of biological networks. Bioinformatics 24: 282–284 - PubMed
- CNHLPP Consortium (2010) First insight into the human liver proteome from PROTEOME(SKY)-LIVER(Hu) 1.0, a publicly available database. J Proteome Res 9: 79–94 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases