A draft genome of Yersinia pestis from victims of the Black Death - PubMed (original) (raw)
. 2011 Oct 12;478(7370):506-10.
doi: 10.1038/nature10549.
Verena J Schuenemann, G Brian Golding, Hernán A Burbano, Nicholas Waglechner, Brian K Coombes, Joseph B McPhee, Sharon N DeWitte, Matthias Meyer, Sarah Schmedes, James Wood, David J D Earn, D Ann Herring, Peter Bauer, Hendrik N Poinar, Johannes Krause
Affiliations
- PMID: 21993626
- PMCID: PMC3690193
- DOI: 10.1038/nature10549
A draft genome of Yersinia pestis from victims of the Black Death
Kirsten I Bos et al. Nature. 2011.
Erratum in
- Nature. 2011 Dec 8;480(7376):278
Abstract
Technological advances in DNA recovery and sequencing have drastically expanded the scope of genetic analyses of ancient specimens to the extent that full genomic investigations are now feasible and are quickly becoming standard. This trend has important implications for infectious disease research because genomic data from ancient microbes may help to elucidate mechanisms of pathogen evolution and adaptation for emerging and re-emerging infections. Here we report a reconstructed ancient genome of Yersinia pestis at 30-fold average coverage from Black Death victims securely dated to episodes of pestilence-associated mortality in London, England, 1348-1350. Genetic architecture and phylogenetic analysis indicate that the ancient organism is ancestral to most extant strains and sits very close to the ancestral node of all Y. pestis commonly associated with human infection. Temporal estimates suggest that the Black Death of 1347-1351 was the main historical event responsible for the introduction and widespread dissemination of the ancestor to all currently circulating Y. pestis strains pathogenic to humans, and further indicates that contemporary Y. pestis epidemics have their origins in the medieval era. Comparisons against modern genomes reveal no unique derived positions in the medieval organism, indicating that the perceived increased virulence of the disease during the Black Death may not have been due to bacterial phenotype. These findings support the notion that factors other than microbial genetics, such as environment, vector dynamics and host susceptibility, should be at the forefront of epidemiological discussions regarding emerging Y. pestis infections.
Figures
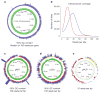
Figure 1. Coverage plots for genomic regions sequenced
a, c–e, Coverage plots for the chromosome (a) and the plasmids pMT1 (c), pCD1 (d) and pPCP (e). Coverage in blue, GC content in green. Scale lines indicate 10-, 20-, 30-, 40- and 50-fold coverage and 10%, 20%, 30%, 40% and 50% GC content. For plasmids, red corresponds to coding regions, yellow to mobile elements. Chromosome shows median coverage per gene. Plasmids show each site plotted. Coverage distributions for the plasmids are shown in Supplementary Fig. 5. b, Distributions show chromosomal coverage of array 1 (blue) and array 2 (red), indicating that deeper sequencing increases the number of reads per site, but does not substantially influence overall coverage.
Figure 2. Alignment of mapped reconstructed contigs against CO92 and Microtus genomes
Reads mapped at positions A (blue) and B (green) are 231 kb apart in the linearized CO92 genome. Adjacent sequence is high coverage although only 18× and 20× is shown due to space constraints (black) for A and B, respectively. The structural variant was assembled using reads that did not map to CO92 (red). Its position is shown on the linearized Microtus 91001 chromosome where the 9,096 bp contig maps with 100% identity.
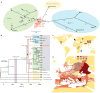
Figure 3. Phylogenetic placement and historical context for the East Smithfield strain
a, Median network of ancient and modern Y. pestis based on 1,694 variant positions in modern genomes. Coloured circles represent different clades as defined in ref. . Gray circles represent hypothetical nodes. b, Phylogenetic tree using 1,694 variable positions. Divergence time intervals are shown in calendar years, with neighbour-joining bootstrap support (blue italic) and Bayesian posterior probability (blue). Grey box indicates known human pathogenic strains. A, NZ ACNQ01000; Nepal516, NC 008149; KIM10, NC 004088; B, NZ AAYT01000; C, NZ ABAT01000; D, NZ ACNS01000; E, NZ AAYS01000; F, NZ AAOS02000; CO92, NC 003143; G, NZ ABCD01000; H, NZ AAYV01000; I, NC 014029; J, NZ AAYR01000; Antiqua, NC 008150. c, Geographical origin of genome sequences used in a and b. d, Geographical spread of the Black Death from infection routes reported in ref. .
Comment in
- Genomics: Plague's progress.
Holmes EC. Holmes EC. Nature. 2011 Oct 26;478(7370):465-6. doi: 10.1038/478465a. Nature. 2011. PMID: 22031436 No abstract available.
Similar articles
- Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death.
Schuenemann VJ, Bos K, DeWitte S, Schmedes S, Jamieson J, Mittnik A, Forrest S, Coombes BK, Wood JW, Earn DJ, White W, Krause J, Poinar HN. Schuenemann VJ, et al. Proc Natl Acad Sci U S A. 2011 Sep 20;108(38):E746-52. doi: 10.1073/pnas.1105107108. Epub 2011 Aug 29. Proc Natl Acad Sci U S A. 2011. PMID: 21876176 Free PMC article. - Comparative scaffolding and gap filling of ancient bacterial genomes applied to two ancient Yersinia pestis genomes.
Luhmann N, Doerr D, Chauve C. Luhmann N, et al. Microb Genom. 2017 Jul 8;3(9):e000123. doi: 10.1099/mgen.0.000123. eCollection 2017 Sep. Microb Genom. 2017. PMID: 29114402 Free PMC article. - Plague genome: The Black Death decoded.
Callaway E. Callaway E. Nature. 2011 Oct 25;478(7370):444-6. doi: 10.1038/478444a. Nature. 2011. PMID: 22031418 No abstract available. - Genome and Evolution of Yersinia pestis.
Cui Y, Song Y. Cui Y, et al. Adv Exp Med Biol. 2016;918:171-192. doi: 10.1007/978-94-024-0890-4_6. Adv Exp Med Biol. 2016. PMID: 27722863 Review. - Plague in the genomic area.
Drancourt M. Drancourt M. Clin Microbiol Infect. 2012 Mar;18(3):224-30. doi: 10.1111/j.1469-0691.2012.03774.x. Clin Microbiol Infect. 2012. PMID: 22369155 Review.
Cited by
- Pervasive findings of directional selection realize the promise of ancient DNA to elucidate human adaptation.
Akbari A, Barton AR, Gazal S, Li Z, Kariminejad M, Perry A, Zeng Y, Mittnik A, Patterson N, Mah M, Zhou X, Price AL, Lander ES, Pinhasi R, Rohland N, Mallick S, Reich D. Akbari A, et al. bioRxiv [Preprint]. 2024 Sep 15:2024.09.14.613021. doi: 10.1101/2024.09.14.613021. bioRxiv. 2024. PMID: 39314480 Free PMC article. Preprint. - Dying of pestilence: Stature and mortality from the Black Death in 14th-century Kyrgyzstan.
Hansen DW, DeWitte SN, Slavin P. Hansen DW, et al. Am J Biol Anthropol. 2024 Nov;185(3):e25009. doi: 10.1002/ajpa.25009. Epub 2024 Sep 5. Am J Biol Anthropol. 2024. PMID: 39238322 - Differential pathogenicity and lethality of bubonic plague (1720-1945) by sex, age and place.
Mongillo J, Zedda N, Rinaldo N, Bellini T, Manfrinato MC, Du Z, Yang R, Stenseth NC, Bramanti B. Mongillo J, et al. Proc Biol Sci. 2024 Aug;291(2027):20240724. doi: 10.1098/rspb.2024.0724. Epub 2024 Jul 24. Proc Biol Sci. 2024. PMID: 39045692 Free PMC article. - Sequential trypsin and ProAlanase digestions unearth immunological protein biomarkers shrouded by skeletal collagen.
Wilkin S, Lanigan LT, Montes N, Sharma M, Avanzi C, Sejdiu D, Majander K, Pfrengle S, Chiang Y, Kunz L, Dittmann A, Rühli F, Singh P, Coll MF, Collins MJ, Taurozzi AJ, Schuenemann VJ. Wilkin S, et al. iScience. 2024 Apr 3;27(5):109663. doi: 10.1016/j.isci.2024.109663. eCollection 2024 May 17. iScience. 2024. PMID: 38655200 Free PMC article. - Identification of microbial pathogens in Neolithic Scandinavian humans.
Bergfeldt N, Kırdök E, Oskolkov N, Mirabello C, Unneberg P, Malmström H, Fraser M, Sanchez-Quinto F, Jorgensen R, Skar B, Lidén K, Jakobsson M, Storå J, Götherström A. Bergfeldt N, et al. Sci Rep. 2024 Mar 7;14(1):5630. doi: 10.1038/s41598-024-56096-0. Sci Rep. 2024. PMID: 38453993 Free PMC article.
References
- Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nature Rev Genet. 2011;12:603–614. - PubMed
- Benedictow OJ. The Black Death 1346–1353: The Complete History. Boydell Press; 2004.
- Scott S, Duncan CJ. Biology of Plagues. Cambridge Univ. Press; 2001.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical