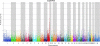Genome-wide association study among four horse breeds identifies a common haplotype associated with in vitro CD3+ T cell susceptibility/resistance to equine arteritis virus infection - PubMed (original) (raw)
Genome-wide association study among four horse breeds identifies a common haplotype associated with in vitro CD3+ T cell susceptibility/resistance to equine arteritis virus infection
Yun Young Go et al. J Virol. 2011 Dec.
Abstract
Previously, we have shown that horses could be divided into susceptible and resistant groups based on an in vitro assay using dual-color flow cytometric analysis of CD3+ T cells infected with equine arteritis virus (EAV). Here, we demonstrate that the differences in in vitro susceptibility of equine CD3+ T lymphocytes to EAV infection have a genetic basis. To investigate the possible hereditary basis for this trait, we conducted a genome-wide association study (GWAS) to compare susceptible and resistant phenotypes. Testing of 267 DNA samples from four horse breeds that had a susceptible or a resistant CD3+ T lymphocyte phenotype using both Illumina Equine SNP50 BeadChip and Sequenom's MassARRAY system identified a common, genetically dominant haplotype associated with the susceptible phenotype in a region of equine chromosome 11 (ECA11), positions 49572804 to 49643932. The presence of a common haplotype indicates that the trait occurred in a common ancestor of all four breeds, suggesting that it may be segregated among other modern horse breeds. Biological pathway analysis revealed several cellular genes within this region of ECA11 encoding proteins associated with virus attachment and entry, cytoskeletal organization, and NF-κB pathways that may be associated with the trait responsible for the in vitro susceptibility/resistance of CD3+ T lymphocytes to EAV infection. The data presented in this study demonstrated a strong association of genetic markers with the trait, representing de facto proof that the trait is under genetic control. To our knowledge, this is the first GWAS of an equine infectious disease and the first GWAS of equine viral arteritis.
Figures
Fig. 1.
Schematic representation of the study design. A total of 310 horses of different breeds, American Saddlebred (ASB; n = 60), Standardbred (STB; n = 60), Thoroughbred (TB; n = 137), and Quarter Horse (QH n = 53), were used in the study. Horses were phenotyped first and grouped into the susceptible or resistant phenotype group based on their CD3+ T cell infectivity to EAV. To identify the region associated with EAV-susceptible/resistant phenotype, gDNA from 37 TB horses was isolated and analyzed using Illumina Equine SNP50. Results from the initial study were confirmed by genotyping 267 additional horses, including TB horses tested with Illumina Equine SNP50, with MassARRAY system technologies.
Fig. 2.
Effect of breeds in prevalence of T cell susceptible/resistant phenotypes. (A) Representative dot plots from flow cytometry analysis for each breed are shown. The mean percentage of CD3+ T cells with intracellular EAV NSP1 antigen is indicated in the right upper quadrant for the susceptible phenotype. (B) The percentages of susceptible (black) and resistant (white) phenotypes for each breed are indicated in a bar graph. Seroprevalences of each breed (red) are indicated below the bar, representing phenotypic prevalence where available. TB, Thoroughbred (n = 137); STB, Standardbred (n = 60); ASB, American Saddlebred (n = 60); QH, Quarter Horse (n = 53).
Fig. 3.
Manhattan plot showing the distribution of probability values (−log10 transformed) for the 42,506 SNPs investigated using the 37 Thoroughbred horses (16 were susceptible and 21 were resistant). Genomic positions are indicated by chromosomes with different colors.
Fig. 4.
Linkage disequilibrium (LD) plots for all breeds, showing the region used for defining the EAV susceptibility haplotype (ECA11 positions 49572804 to 49643932). The haplotype block is highlighted with a black line.
Fig. 5.
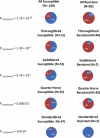
Frequency of the GGGGAGGT haplotype found for selected SNPs between ECA11 positions 49572804 to 49643932 among horses susceptible and resistant for the EAV in vitro infection phenotype. The blue area in the pie chart represents the proportion of the GGGGAGGT haplotype, and the red area represents the allelic haplotypes. The frequency represented by each section of the pie chart is shown on the pie chart in white. Data are represented for all horses together (All) and for the individual breeds (Thoroughbred, American Saddlebred, Quarter Horse, and Standardbred). The statistical significance for the frequency differences of the GGGGAGGT haplotype between susceptible and resistant horses is shown to the left of each set of pie charts (P“GGGGAGGT”).
Fig. 6.
(A) Three top networks retrieved using unsupervised IPA were merged into a single interaction network. Red nodes represent molecules encoded by genes found in the region of ECA11; white nodes are genes identified by IPA with direct or indirect interactions with genes of ECA11. The blue nodes represent molecules/complexes identified by IPA involved in canonical pathways. Classification of candidate molecules based on their molecular functions (B) and biological processes (C).
Fig. 7.
Subcellular location of candidate genes and EAV life cycle. The function and subcellular location of candidate molecules were manually curated based on PANTHER, IPA, and published literature search. Subsequently, molecules were mapped along with stages of the EAV life cycle at the position most likely to interact with the virus life cycle. Green circles, molecules encoded by genes located in the haplotype block; red circles, putative molecules encoded by genes found within 500 kb upstream and downstream of the haplotype block; blue circles, molecules in the vicinity interacting with genes located in the region retrieved in the network analysis; black circles, viral proteins.
Similar articles
- Allelic Variation in CXCL16 Determines CD3+ T Lymphocyte Susceptibility to Equine Arteritis Virus Infection and Establishment of Long-Term Carrier State in the Stallion.
Sarkar S, Bailey E, Go YY, Cook RF, Kalbfleisch T, Eberth J, Chelvarajan RL, Shuck KM, Artiushin S, Timoney PJ, Balasuriya UB. Sarkar S, et al. PLoS Genet. 2016 Dec 8;12(12):e1006467. doi: 10.1371/journal.pgen.1006467. eCollection 2016 Dec. PLoS Genet. 2016. PMID: 27930647 Free PMC article. - Assessment of correlation between in vitro CD3+ T cell susceptibility to EAV infection and clinical outcome following experimental infection.
Go YY, Cook RF, Fulgêncio JQ, Campos JR, Henney P, Timoney PJ, Horohov DW, Balasuriya UB. Go YY, et al. Vet Microbiol. 2012 May 25;157(1-2):220-5. doi: 10.1016/j.vetmic.2011.11.031. Epub 2011 Dec 2. Vet Microbiol. 2012. PMID: 22177968 - Evidence that in vitro susceptibility of CD3+ T lymphocytes to equine arteritis virus infection reflects genetic predisposition of naturally infected stallions to become carriers of the virus.
Go YY, Bailey E, Timoney PJ, Shuck KM, Balasuriya UB. Go YY, et al. J Virol. 2012 Nov;86(22):12407-10. doi: 10.1128/JVI.01698-12. Epub 2012 Aug 29. J Virol. 2012. PMID: 22933293 Free PMC article. - Equine arteritis virus.
Balasuriya UB, Go YY, MacLachlan NJ. Balasuriya UB, et al. Vet Microbiol. 2013 Nov 29;167(1-2):93-122. doi: 10.1016/j.vetmic.2013.06.015. Epub 2013 Jul 3. Vet Microbiol. 2013. PMID: 23891306 Free PMC article. Review. - A perspective on equine viral arteritis (infectious arteritis of horses).
Timoney PJ, Klingeborn B, Lucas MH. Timoney PJ, et al. Rev Sci Tech. 1996 Sep;15(3):1203-8. doi: 10.20506/rst.15.3.971. Rev Sci Tech. 1996. PMID: 9025155 Review. No abstract available.
Cited by
- Development of a TaqMan® Allelic Discrimination qPCR Assay for Rapid Detection of Equine CXCL16 Allelic Variants Associated With the Establishment of Long-Term Equine Arteritis Virus Carrier State in Stallions.
Thieulent CJ, Carossino M, Balasuriya UBR, Graves K, Bailey E, Eberth J, Canisso IF, Andrews FM, Keowen ML, Go YY. Thieulent CJ, et al. Front Genet. 2022 Apr 13;13:871875. doi: 10.3389/fgene.2022.871875. eCollection 2022. Front Genet. 2022. PMID: 35495124 Free PMC article. - Genome-Wide Association Study Using Individual Single-Nucleotide Polymorphisms and Haplotypes for Erythrocyte Traits in Alpine Merino Sheep.
Zhu S, Guo T, Zhao H, Qiao G, Han M, Liu J, Yuan C, Wang T, Li F, Yue Y, Yang B. Zhu S, et al. Front Genet. 2020 Jul 31;11:848. doi: 10.3389/fgene.2020.00848. eCollection 2020. Front Genet. 2020. PMID: 32849829 Free PMC article. - Host Transcriptional Response to Persistent Infection with a Live-Attenuated Porcine Reproductive and Respiratory Syndrome Virus Strain.
Chaudhari J, Liew CS, Workman AM, Riethoven JM, Steffen D, Sillman S, Vu HLX. Chaudhari J, et al. Viruses. 2020 Jul 28;12(8):817. doi: 10.3390/v12080817. Viruses. 2020. PMID: 32731586 Free PMC article. - Equine arteritis virus long-term persistence is orchestrated by CD8+ T lymphocyte transcription factors, inhibitory receptors, and the CXCL16/CXCR6 axis.
Carossino M, Dini P, Kalbfleisch TS, Loynachan AT, Canisso IF, Cook RF, Timoney PJ, Balasuriya UBR. Carossino M, et al. PLoS Pathog. 2019 Jul 29;15(7):e1007950. doi: 10.1371/journal.ppat.1007950. eCollection 2019 Jul. PLoS Pathog. 2019. PMID: 31356622 Free PMC article. - Intrahost Selection Pressure Drives Equine Arteritis Virus Evolution during Persistent Infection in the Stallion Reproductive Tract.
Nam B, Mekuria Z, Carossino M, Li G, Zheng Y, Zhang J, Cook RF, Shuck KM, Campos JR, Squires EL, Troedsson MHT, Timoney PJ, Balasuriya UBR. Nam B, et al. J Virol. 2019 May 29;93(12):e00045-19. doi: 10.1128/JVI.00045-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30918077 Free PMC article.
References
- Abel S., et al. 2004. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J. Immunol. 172:6362–6372 - PubMed
- Ait-Ali T., et al. 2009. Functional analysis of the porcine USP18 and its role during porcine arterivirus replication. Gene 439:35–42 - PubMed
- Balasuriya U. B., MacLachlan N. J. 2007. Equine viral arteritis, p. 153–164 In Sellon D. C., Long M. T.(ed.), Equine infectious diseases. Elsevier, St. Louis, MO
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources