A genome-wide survey for SNPs altering microRNA seed sites identifies functional candidates in GWAS - PubMed (original) (raw)
A genome-wide survey for SNPs altering microRNA seed sites identifies functional candidates in GWAS
Kris Richardson et al. BMC Genomics. 2011.
Abstract
Background: Gene variants within regulatory regions are thought to be major contributors of the variation of complex traits/diseases. Genome wide association studies (GWAS), have identified scores of genetic variants that appear to contribute to human disease risk. However, most of these variants do not appear to be functional. Thus, the significance of the association may be brought up by still unknown mechanisms or by linkage disequilibrium (LD) with functional polymorphisms. In the present study, focused on functional variants related with the binding of microRNAs (miR), we utilized SNP data, including newly released 1000 Genomes Project data to perform a genome-wide scan of SNPs that abrogate or create miR recognition element (MRE) seed sites (MRESS).
Results: We identified 2723 SNPs disrupting, and 22295 SNPs creating MRESSs. We estimated the percent of SNPs falling within both validated (5%) and predicted conserved MRESSs (3%). We determined 87 of these MRESS SNPs were listed in GWAS association studies, or in strong LD with a GWAS SNP, and may represent the functional variants of identified GWAS SNPs. Furthermore, 39 of these have evidence of co-expression of target mRNA and the predicted miR. We also gathered previously published eQTL data supporting a functional role for four of these SNPs shown to associate with disease phenotypes. Comparison of FST statistics (a measure of population subdivision) for predicted MRESS SNPs against non MRESS SNPs revealed a significantly higher (P = 0.0004) degree of subdivision among MRESS SNPs, suggesting a role for these SNPs in environmentally driven selection.
Conclusions: We have demonstrated the potential of publicly available resources to identify high priority candidate SNPs for functional studies and for disease risk prediction.
Figures
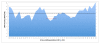
Figure 1
A measure of SNP density (SNPs/kb) generated from the analysis of a 6 base window sliding over a 42-base region - centered on the first position of the seed site - of 606 validated MREs. The black line indicates the number of SNPs/kB across the 42 base region. The red line indicates the average SNP density across the 6 windows of seed positions 2-7.
Figure 2
Four SNPs found to associate, or be in LD with a SNP that associates, with a trait(s) relevant to disease. Each panel depicts the mRNA-miR interaction and the effect of the SNP on this interaction. Plots were generated using the Genvar web tool and published expression data from Nica, et al. rho = correlation between genotype and transcript levels. p = t = test statistic for correlation. padj = adjusted pvalue for correlation. F = -Fat cell biopsy (n = 160), L = LCL cells (n = 166), and S = skin cell biopsy (n = 160). Twin1 = Unrelated twin group 1. Twin2 = unrelated twin group 2.
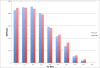
Figure 3
A plot showing the number of combined MRESS and CNM SNPs or non MRESS and non CNM 3'UTR SNPs across 10 FST bins. Data plotted compares 2448 MRESS and CNM SNPs with FST data and a random sample of 2448 FST values from the remainder of 3'UTR SNPs. A significant difference between mean FST values for combined MRESS and CNM SNPs and FST values for the remaining 3'UTR SNPs was observed (P = 0.0004).
Similar articles
- Genome-wide association study combined with biological context can reveal more disease-related SNPs altering microRNA target seed sites.
Wu D, Yang G, Zhang L, Xue J, Wen Z, Li M. Wu D, et al. BMC Genomics. 2014 Aug 8;15(1):669. doi: 10.1186/1471-2164-15-669. BMC Genomics. 2014. PMID: 25106527 Free PMC article. - MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs.
Liu C, Zhang F, Li T, Lu M, Wang L, Yue W, Zhang D. Liu C, et al. BMC Genomics. 2012 Nov 23;13:661. doi: 10.1186/1471-2164-13-661. BMC Genomics. 2012. PMID: 23173617 Free PMC article. - In silico prioritisation of microRNA-associated common variants in multiple sclerosis.
Fashina IA, McCoy CE, Furney SJ. Fashina IA, et al. Hum Genomics. 2023 Mar 30;17(1):31. doi: 10.1186/s40246-023-00478-4. Hum Genomics. 2023. PMID: 36991503 Free PMC article. - Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation.
Croteau-Chonka DC, Rogers AJ, Raj T, McGeachie MJ, Qiu W, Ziniti JP, Stubbs BJ, Liang L, Martinez FD, Strunk RC, Lemanske RF Jr, Liu AH, Stranger BE, Carey VJ, Raby BA. Croteau-Chonka DC, et al. PLoS One. 2015 Oct 16;10(10):e0140758. doi: 10.1371/journal.pone.0140758. eCollection 2015. PLoS One. 2015. PMID: 26474488 Free PMC article. Review. - Critical Analysis of Genome-Wide Association Studies: Triple Negative Breast Cancer Quae Exempli Causa.
Jurj MA, Buse M, Zimta AA, Paradiso A, Korban SS, Pop LA, Berindan-Neagoe I. Jurj MA, et al. Int J Mol Sci. 2020 Aug 14;21(16):5835. doi: 10.3390/ijms21165835. Int J Mol Sci. 2020. PMID: 32823908 Free PMC article. Review.
Cited by
- Effects of genetic variations on microRNA: target interactions.
Liu C, Rennie WA, Carmack CS, Kanoria S, Cheng J, Lu J, Ding Y. Liu C, et al. Nucleic Acids Res. 2014 Sep;42(15):9543-52. doi: 10.1093/nar/gku675. Epub 2014 Jul 31. Nucleic Acids Res. 2014. PMID: 25081214 Free PMC article. - regQTLs: Single nucleotide polymorphisms that modulate microRNA regulation of gene expression in tumors.
Wilk G, Braun R. Wilk G, et al. PLoS Genet. 2018 Dec 17;14(12):e1007837. doi: 10.1371/journal.pgen.1007837. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30557297 Free PMC article. - An update on the roles of transcription factor Ets1 in autoimmune diseases.
Garrett-Sinha LA. Garrett-Sinha LA. WIREs Mech Dis. 2023 Nov-Dec;15(6):e1627. doi: 10.1002/wsbm.1627. Epub 2023 Aug 10. WIREs Mech Dis. 2023. PMID: 37565573 Free PMC article. Review. - How Gene Networks Can Uncover Novel CVD Players.
Parnell LD, Casas-Agustench P, Iyer LK, Ordovas JM. Parnell LD, et al. Curr Cardiovasc Risk Rep. 2014 Jan;8:372. doi: 10.1007/s12170-013-0372-3. Curr Cardiovasc Risk Rep. 2014. PMID: 24683432 Free PMC article. - Applications of the 1000 Genomes Project resources.
Zheng-Bradley X, Flicek P. Zheng-Bradley X, et al. Brief Funct Genomics. 2017 May 1;16(3):163-170. doi: 10.1093/bfgp/elw027. Brief Funct Genomics. 2017. PMID: 27436001 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous