Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge - PubMed (original) (raw)
Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge
Tong Zhang et al. PLoS One. 2011.
Abstract
The overuse or misuse of antibiotics has accelerated antibiotic resistance, creating a major challenge for the public health in the world. Sewage treatment plants (STPs) are considered as important reservoirs for antibiotic resistance genes (ARGs) and activated sludge characterized with high microbial density and diversity facilitates ARG horizontal gene transfer (HGT) via mobile genetic elements (MGEs). However, little is known regarding the pool of ARGs and MGEs in sludge microbiome. In this study, the transposon aided capture (TRACA) system was employed to isolate novel plasmids from activated sludge of one STP in Hong Kong, China. We also used Illumina Hiseq 2000 high-throughput sequencing and metagenomics analysis to investigate the plasmid metagenome. Two novel plasmids were acquired from the sludge microbiome by using TRACA system and one novel plasmid was identified through metagenomics analysis. Our results revealed high levels of various ARGs as well as MGEs for HGT, including integrons, transposons and plasmids. The application of the TRACA system to isolate novel plasmids from the environmental metagenome, coupled with subsequent high-throughput sequencing and metagenomic analysis, highlighted the prevalence of ARGs and MGEs in microbial community of STPs.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
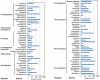
Figure 1. Abundances of bacterial hosts of plasmids in activated sludge of Shatin STP.
Figure 2. Open reading frames (ORFs) on the three plasmids detected in activated sludge of Shatin STP. pST1 structure was identified by assembling the sequencing reads using SOAPdenovo (BGI, Shenzhen, China).
pST2 and pST10 were cloned with culture-independent transposon aided capture system and DNA sequencing were conducted applying the primer walking strategy.
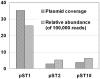
Figure 3. Relative abundances of the three plasmids in activated sludge of Shatin STP.
The relative abundance is given in relation to the total number of sequencing reads and the plasmid coverage was estimated by counting the number of reads aligned at each base.
Figure 4. Relative abundances of various ARGs in activated sludge of Shatin STP.
The relative abundance is given in relation to the total number of ARG-matched reads.
Similar articles
- Fate of antibiotic resistant E. coli and antibiotic resistance genes during full scale conventional and advanced anaerobic digestion of sewage sludge.
Redhead S, Nieuwland J, Esteves S, Lee DH, Kim DW, Mathias J, Cha CJ, Toleman M, Dinsdale R, Guwy A, Hayhurst E. Redhead S, et al. PLoS One. 2020 Dec 1;15(12):e0237283. doi: 10.1371/journal.pone.0237283. eCollection 2020. PLoS One. 2020. PMID: 33259486 Free PMC article. - Shotgun metagenomic analysis reveals the prevalence of antibiotic resistance genes and mobile genetic elements in full scale hospital wastewater treatment plants.
Manoharan RK, Srinivasan S, Shanmugam G, Ahn YH. Manoharan RK, et al. J Environ Manage. 2021 Oct 15;296:113270. doi: 10.1016/j.jenvman.2021.113270. Epub 2021 Jul 14. J Environ Manage. 2021. PMID: 34271348 - The equine hindgut as a reservoir of mobile genetic elements and antimicrobial resistance genes.
Mitchell S, Bull M, Muscatello G, Chapman B, Coleman NV. Mitchell S, et al. Crit Rev Microbiol. 2021 Sep;47(5):543-561. doi: 10.1080/1040841X.2021.1907301. Epub 2021 Apr 24. Crit Rev Microbiol. 2021. PMID: 33899656 Review. - Microbial evolution through horizontal gene transfer by mobile genetic elements.
Tokuda M, Shintani M. Tokuda M, et al. Microb Biotechnol. 2024 Jan;17(1):e14408. doi: 10.1111/1751-7915.14408. Epub 2024 Jan 16. Microb Biotechnol. 2024. PMID: 38226780 Free PMC article. Review.
Cited by
- Isolation, molecular identification, and genomic analysis of Mangrovibacter phragmitis strain ASIOC01 from activated sludge harboring the bioremediation prowess of glycerol and organic pollutants in high-salinity.
Chin HS, Ravi Varadharajulu N, Lin ZH, Chen WY, Zhang ZH, Arumugam S, Lai CY, Yu SS. Chin HS, et al. Front Microbiol. 2024 Jun 25;15:1415723. doi: 10.3389/fmicb.2024.1415723. eCollection 2024. Front Microbiol. 2024. PMID: 38983623 Free PMC article. - Intra- and interpopulation transposition of mobile genetic elements driven by antibiotic selection.
Yao Y, Maddamsetti R, Weiss A, Ha Y, Wang T, Wang S, You L. Yao Y, et al. Nat Ecol Evol. 2022 May;6(5):555-564. doi: 10.1038/s41559-022-01705-2. Epub 2022 Mar 28. Nat Ecol Evol. 2022. PMID: 35347261 - The Resistome of ESKAPEE Pathogens in Untreated and Treated Wastewater: A Polish Case Study.
Hubeny J, Korzeniewska E, Ciesielski S, Płaza G, Harnisz M. Hubeny J, et al. Biomolecules. 2022 Aug 21;12(8):1160. doi: 10.3390/biom12081160. Biomolecules. 2022. PMID: 36009054 Free PMC article. - Plasmid Detection, Characterization, and Ecology.
Smalla K, Jechalke S, Top EM. Smalla K, et al. Microbiol Spectr. 2015 Feb;3(1):PLAS-0038-2014. doi: 10.1128/microbiolspec.PLAS-0038-2014. Microbiol Spectr. 2015. PMID: 26104560 Free PMC article. Review. - Emergence of Carbapenemase Genes in Gram-Negative Bacteria Isolated from the Wastewater Treatment Plant in A Coruña, Spain.
Nasser-Ali M, Aja-Macaya P, Conde-Pérez K, Trigo-Tasende N, Rumbo-Feal S, Fernández-González A, Bou G, Poza M, Vallejo JA. Nasser-Ali M, et al. Antibiotics (Basel). 2024 Feb 17;13(2):194. doi: 10.3390/antibiotics13020194. Antibiotics (Basel). 2024. PMID: 38391580 Free PMC article.
References
- Zhang XX, Zhang T, Fang HHP. Antibiotic resistance genes in water environment. Appl Microbiol Biotechnol. 2009;82:397–414. - PubMed
- Szczepanowski R, Krahn I, Linke B, Goesmann A, Pühler A, et al. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology. 2004;150:3613–30. - PubMed
- Tennstedt T, Szczepanowski R, Krahn K, Pühler A, Schlüter A. Sequence of the 68,869 bp IncP-1α plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid. 2005;53:218–238. - PubMed
- da Silva MF, Vaz-Moreira I, Gonzalez-Pajuelo M, Nunes OC, Manaia CM. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol Ecol. 2007;60:166–176. - PubMed
- Taviani E, Ceccarelli D, Lazaro N, Bani S, Cappuccinelli P, met al. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol Ecol. 2008;64:45–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical